Figure 2. A common core program of regulatory gene expression change in mouse and human.
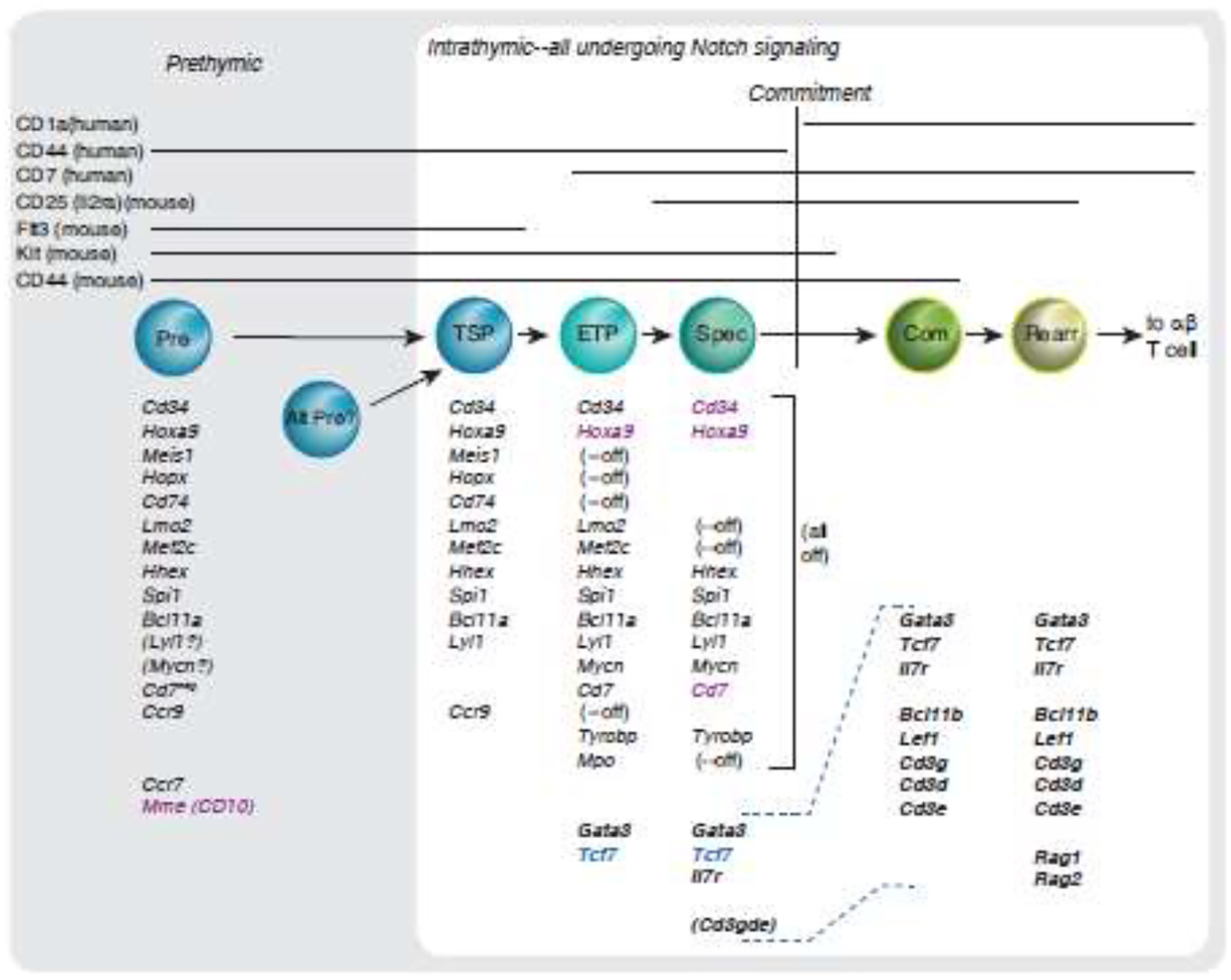
A consensus conserved gene expression program is shared by early T cells, in postnatal mice and humans. The figure shows key regulatory and signaling genes expressed at the corresponding indicated stages in both mouse and human [43–45]. The framework of stages shown is rooted in the TSP1 precursor trajectory with early intrathymic upregulation of Cd7 [44, 45] and the mouse stage equivalents. Stage markers are shown at the top and aligned as in Table 1. Note that mouse and human marker expression patterns are slightly different relative to underlying transcriptomes even when the same marker is tracked in both. Where possible, each gene in a group is listed in the same row across the figure. (--off), silencing of the gene in this row expressed in previous stages. Gene names in bold: T-cell program genes activated in T-lineage precursors by thymic microenvironment. Gene names in black: genes expressed in common between mouse and human cells at corresponding stages. Gene names in magenta: genes expressed in indicated stages persist later than in mouse, or lack a mouse equivalent. Gene name in blue: gene expressed at this stage is activated earlier than in human lineage. (Cd3gde), slight upregulation of the Cd3g, Cd3d, Cd3e gene cluster detectable before commitment, although greatly enhanced after commitment. (Lyl1), (Mycn) in Prethymic: probable expression but not reported directly. Evidence for gene expression in the prethymic mouse precursor is based on LMPP phenotypes and evidence from genetic studies of Ccr9 and Ccr7 impact on thymic immigration [98, 99]. Note that these lists are approximate, not comprehensive, and quantitative changes in expression are not represented. Patterns are gleaned from data shown in the figures and tables of the references as published [43–45] and not from reanalysis of all genomic datasets with consistent statistical criteria; comparative levels of genes shown were not always available across all populations. Population abbreviations: TSP, stage 0 in Table 1. ETP, stage 1. Spec, specified (stage 2). Com, committed (stage 3). Rearr, TCR gene rearranging (stage 4). Dashed lines in gene lists from Spec to Com aid to align gene groups.
