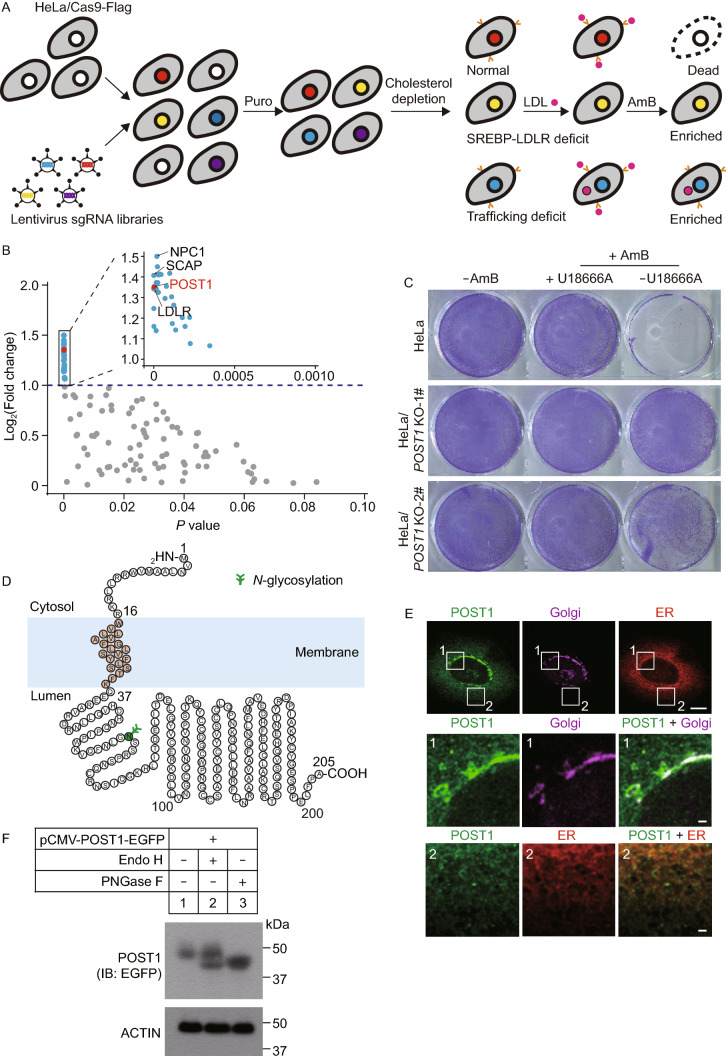
Figure 1.
Genome-wide screen identifies that POST1 is involved in cholesterol metabolism. (A) Schematic representation of the screening strategy. HeLa cells stably expressing Cas9-Flag were transduced with lentivirus expressing a genome-wide sgRNA library and then treated with puromycin (Puro) for 4 days. Surviving cells were depleted of cholesterol by incubating in the medium containing 5% lipoprotein-deficient serum (LPDS) plus 10 µmol/L mevalonate and 1 µmol/L lovastatin for 16 h. Cells were then incubated with 50 µg/mL LDL for 4 h followed by 300 µg/mL amphotericin B (AmB) for 1 h. AmB could bind PM cholesterol, form pores and kill normal cells. The mutant cells defective in the SREBP-LDLR axis or cholesterol trafficking were resistant to AmB because of less PM cholesterol. After five rounds of challenges, the sgRNA inserts from surviving cells and those from transduced cells prior to the first challenge were amplified and subjected to deep sequencing. (B) Scatter plot showing 115 highly enriched genes (Supplementary Material, Table S1) in (A). Genes with a phenotype value (fold change [log2]) >1 and P-value < 0.001 are in blue (except for POST1 in red) and are shown in smaller scales of x- and y-axes (inset). Those with a phenotype value <1 are in gray. (C) HeLa cells and two lines of POST1 KO cells generated by the CRISPR/Cas9 technique (POST1 KO-1# and POST1 KO-2#) were depleted of cholesterol for 16 h. Cells were then incubated in the medium containing 5% LPDS, 50 µg/mL LDL and 1 µmol/L lovastatin in the absence or presence of 2 µg/mL U18666A for 4 h, and then in 300 µg/mL AmB for 1 h. (D) The predicted topology of human POST1 protein. (E) HeLa cells were transfected with pCMV-POST1-EGFP (green) and pCMV-DsRed2-KDEL (red) for 48 h, and immunostained with the antibody against GM130 (magenta). Boxed areas are shown at a higher magnification as numbered below. Scale bar, 10 µm (main), 1 µm (inset). (F) HeLa cells were transfected with pCMV-POST1-EGFP for 48 h and harvested. Lysates were treated with 10 units/μL Endo H or 5 units/μL PNGase F as indicated prior to immunoblotting