Figure 4. R116 is necessary for viral entry into fibroblasts.
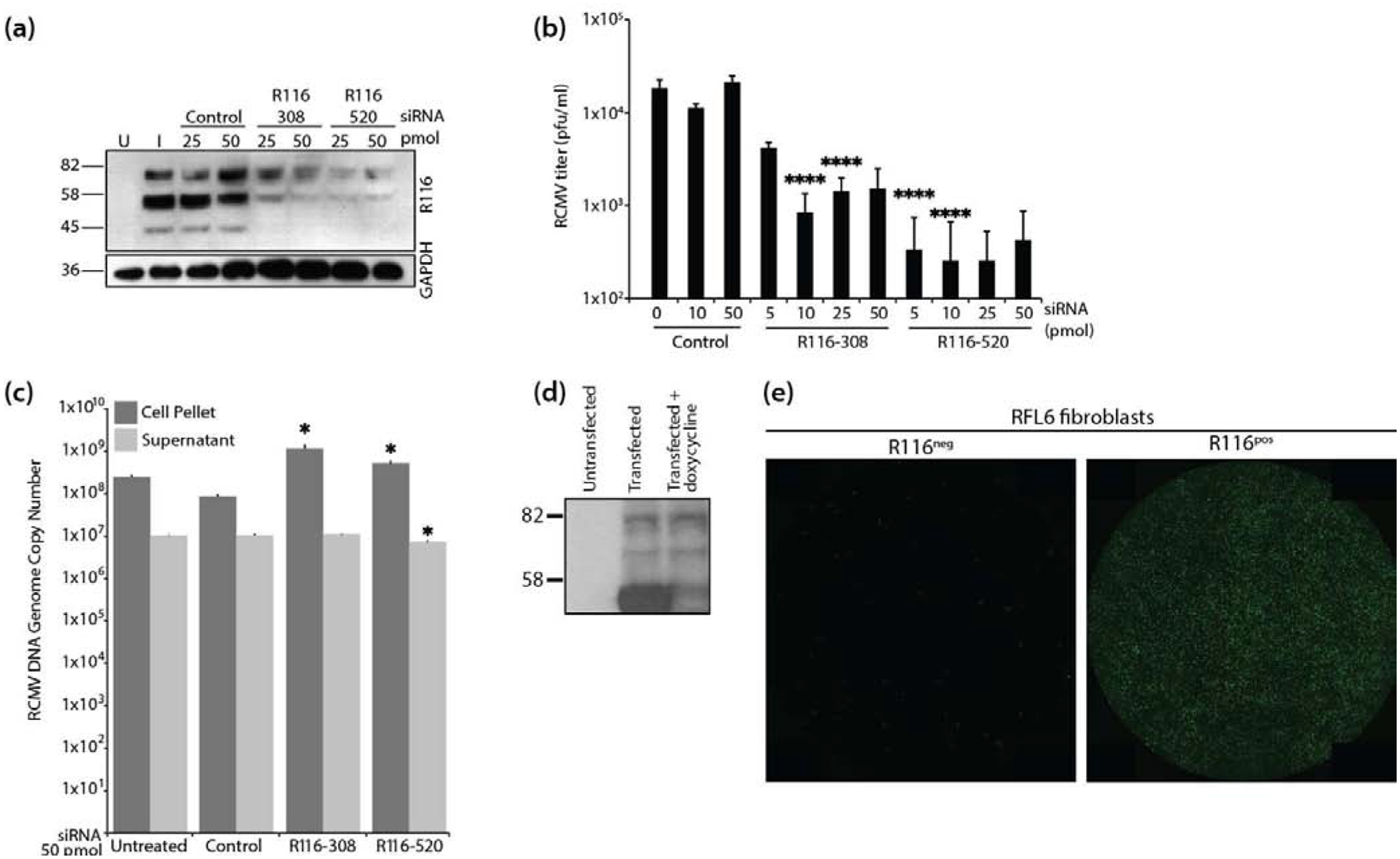
(a-c) In triplicate wells RFL6 fibroblasts were transfected with a control siRNA or siRNAs targeting sequences present with the R116 gene. At 24 hours post transfection, the cells were infected with RCMV (MOI=1.0) and allowed to incubate for 4 hours at which time the virus inocula were removed and the cells were washed twice with fresh medium. At 48 hpi, supernatants and cell pellets were collected and analyzed by: (a) Western blotting for R116 and GAPDH; (b) Plaque assay for the presence of infectious virus. Statistical significance was determined by Student’s t-test [control vs R116 siRNA at 10 and 50pmol ****p<0.001, n=3]; and (c) Quantitative PCR for the presence of viral DNA genomes. Statistical significance was determined by Student’s t-test [control vs. siRNA R116–308 cell pellet p=0.02 and supernatant p=0.33, n=3; control vs. siRNA R116–520 cell pellet and supernatant p=0.01, *p<0.05, n=3]. (d-e) R116 expression by RFL6 cells restores infectivity of ΔR116-RCMV. (d) RFL6 fibroblasts were harvested in lysis buffer containing protease inhibitor, analyzed by SDS-PAGE and probed using the anti-R116 polyclonal antibody. Western blots were visualized by autoradiography. (e) GFP expression 5 days after infection with ΔR116-RCMV in triplicate wells wild-type RFL6 fibroblasts (left) or R116-expressing RFL6 fibroblasts (right). Representative images are shown.
