Figure 5. HCMV UL116 is required for entry into fibroblasts.
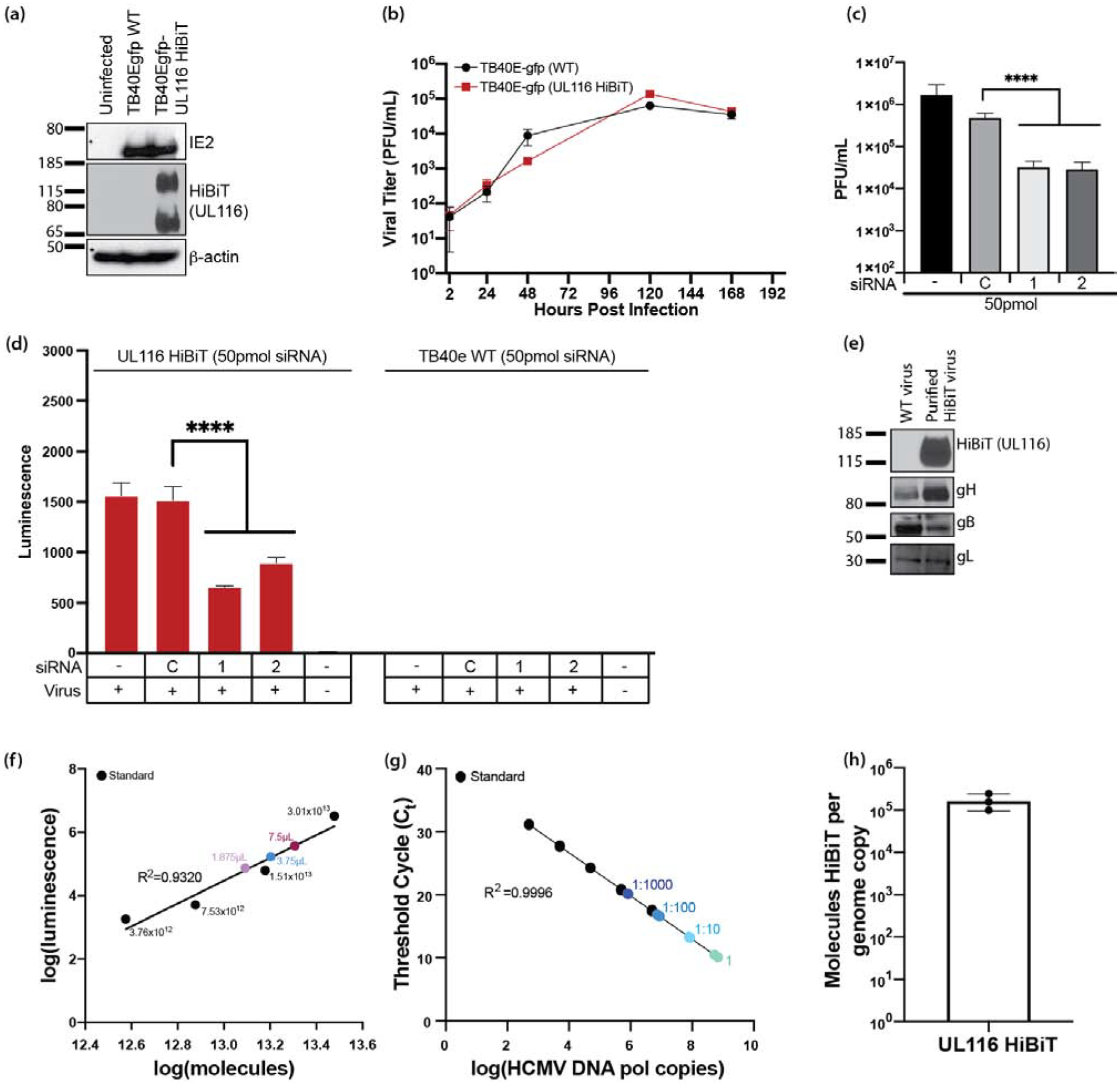
HCMV UL116 was molecularly tagged with the HiBiT tag using BAC recombineering in a TB40e background. (a) NHDF fibroblasts were infected with TB40e GFP HCMV or TB40e GFP UL116-HiBiT HCMV and were harvested in lysis buffer. Western blots were performed for IE2, HiBiT, and β-actin to confirm expression of UL116-HiBiT. (b) NHDF fibroblasts were infected at a MOI=0.3 with HCMV TB40egfp-UL116-HiBiT or WT virus. At 2 hours post infection, the virus inoculums were removed, cells were washed with PBS, and then fresh culture media was added. Supernatants were collected at 24, 48, 120, and 168 hpi and titered using NHDF fibroblasts. (c) NHDF fibroblasts were plated in 24-well plates and treated in quadruplicates. NHDF fibroblasts were transfected with a control siRNA or siRNAs targeting sequences present within the UL116 gene at 50pmol/well. At 24 hours post transfection, the cells were infected with HCMV (MOI=0.3) and incubated for 2 hours at which time the virus inoculums were removed and the cells were washed twice with fresh medium. At 5 days post-infection, supernatants and cell pellets were collected and analyzed by plaque assay for the presence of infectious virus. Statistical significance was determined by one-way ANOVA with Dunnett’s correction for multiple comparisons [****p<0.0001, n=4]. (d) NHDF fibroblasts were seeded in 96-well black plates and transfected with control siRNA or siRNAs targeting sequences present within the UL116 gene. Cells were infected at a MOI=0.3 using a UL116-HiBiT tagged HCMV or TB40e WT HCMV for 2 hours, at which time the virus inoculums were removed and cells were washed with PBS and fresh media was added. At 3 days post-infection cells were harvested and analyzed by HiBiT lytic detection assay. Statistical significance was determined by two-way ANOVA with Dunnett’s correction for multiple comparisons [****p<0.0001]. (e) gH, gB, and gL are incorporated into viral particles of UL116-HiBiT HCMV. WT and UL116-HiBiT HCMV were expanded over NHDF fibroblasts. Viruses were harvested at the time of maximum cytopathic effect and pelleted through a 20% sorbitol cushion. UL116-HiBiT viral particles were further purified over a 10–50% histodenz gradient. Purified viral particles were run on SDS-PAGE gels and western blots for gH, gB, gL, and HiBiT were performed. (f-h) UL116 is incorporated into HCMV virions at 1.06×105 molecules/HCMV genome. HCMV UL116 HiBiT viral particles were isolated over a histodenz gradient and molecules of HiBiT was determined against a standard curve by HiBiT lytic detection assay (f) and genome copies were determined by qPCR for HCMV DNA polymerase (g). HiBiT control protein (Promega) and known genome copies of HCMV TB40e were used as respective assay controls. Ratio of UL116-HiBiT molecules over genome copies was calculated (h).
