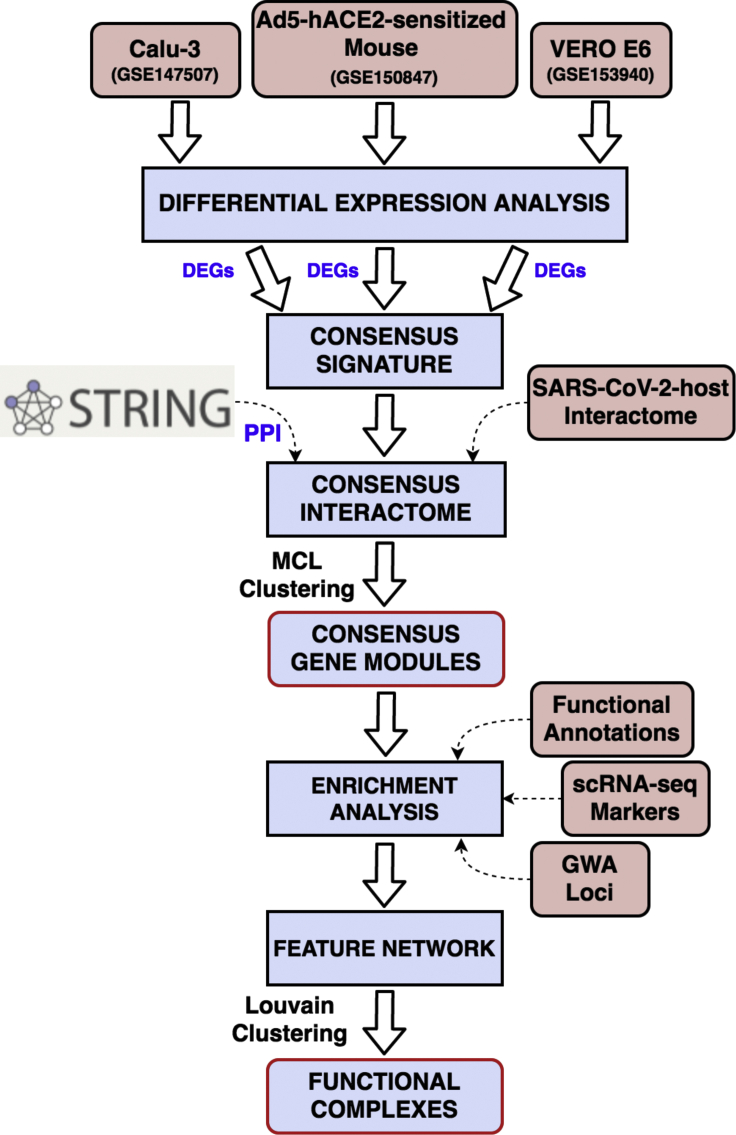
Figure 1.
Schematic representation of the workflow
Transcriptomic data from three SARS-CoV-2 infection models are processed to identify differentially expressed genes (DEGs). Genes that are up- or downregulated in two out of three models are considered as “consensus signature.” The consensus DEGs, along with the SARS-CoV-2-human virus-host interactome, are used to build an integrated network based on known protein-protein interactions (PPIs) retrieved from STRING database (v11). A Markov clustering (MCL) algorithm is then used to identify modules of highly interconnected genes from this integrated interactome. These gene modules are characterized through functional enrichments (pathways, biological processes), lung single-cell markers, and phenotypic trait (genome-wide association [GWA]) loci. In the final step, an enriched feature network is constructed using all the enriched terms and features from the modules to extract functional complexes or communities of highly interconnected features.