Figure 5.
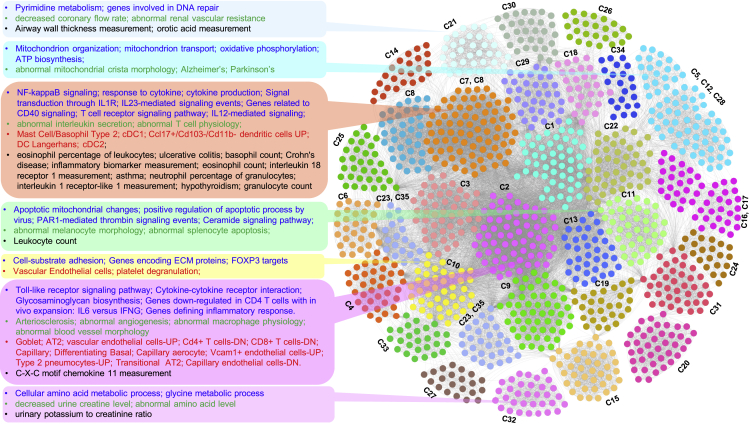
Network visualization of the results from joint analysis of multiple annotations from the 35 gene clusters
Network representation of clustered enriched terms from functional enrichment analysis (multiple annotations such as Gene Ontology, pathways, lung cell types, and GWA loci) of candidate gene clusters from the integrated consensus DEG and SARS-CoV-2-host interacting protein network. Enriched terms from different annotation categories are represented as nodes, while the edges represent shared gene clusters (35 select gene clusters). Representative terms in some of the clustered functional modules are listed on the left side with different font colors representing different annotation categories (blue, biological processes/pathways; green, phenotype; red, cell type; black, GWA trait). The underlying gene clusters (35 select gene clusters) for each of the clustered functional terms are also shown.