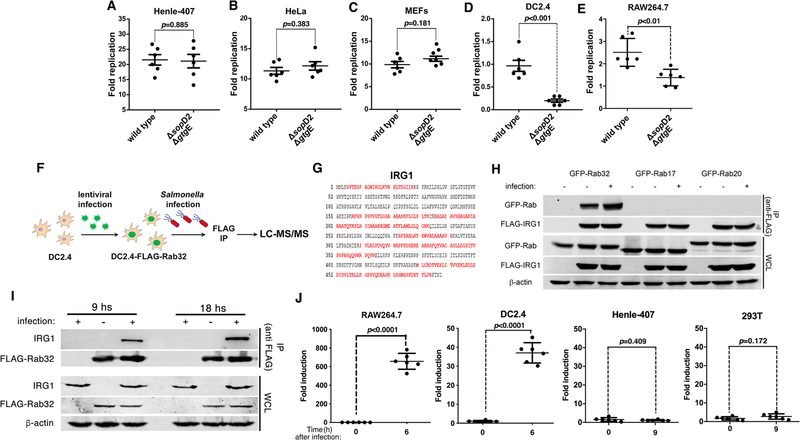
Fig. 1. IRG1 interacts with Rab32 during Salmonella infection.
(A-E) The Rab32-associated pathogen restriction mechanism is manifested in myelocytic but not in epithelial cell lines. The ability of the S. Typhimurium ΔsopD2 ΔgtgE mutant strain to replicate within epithelial (Henle-407 and HeLa) or myelocytic (DC2.4 and RAW264.7) cell lines was evaluated by determining the CFU at different times after infection (MOI = 5). Fold replication represents the difference between the CFU at 1 and 9 hours post infection. Each circle represents the fold replication in each individual determination; the mean ± SEM of all the measurements and the p values of the indicated comparisons (two-sided Student’s t-test) are shown. (F-I) Rab32 interacts with IRG1 after Salmonella infection. DC2.4 cells expressing endogenous levels of FLAG-tagged Rab32 were infected with S. Typhimurium ΔsopD2 ΔgtgE (MOI = 30) and Rab32-interacting proteins were identified by affinity purification and LC—MS/MS analysis (F). The IRG1 peptides identified by the analyses are shown in red (G). (H and I) HEK293T cells transiently co-transfected with a plasmid expressing GFP-tagged Rab32, RAb17, or Rab20, along with a plasmid encoding FLAG-tagged IRG1 (H), or DC2.4 cells stably expressing FLAG-tagged Rab32 (I) were infected with S. Typhimurium ΔgtgE ΔsopD2 for 4 hours (MOI = 5). Cell lysates were then analyzed by immunoprecipitation with anti-FLAG and immunoblotting with anti-GFP, anti-FLAG, anti-IRG1, or anti-β-actin (as loading control) antibodies. IP: immunoprecipitates; WCL: whole-cell lysates. (J) Expression of IRG1 after Salmonella infection. The indicated cell lines were infected with S. Typhimurium ΔsopD2 ΔgtgE mutant strain (MOI = 5) and IRG1 mRNA levels were measured by qPCR 6 or 9 hours after infection. Each circle represents a single determination of the relative levels of IRG1 normalized to the levels of GAPDH; the mean ± SEM of all the measurements and p values of the indicated comparisons (two-sided Student’s t test) are shown.