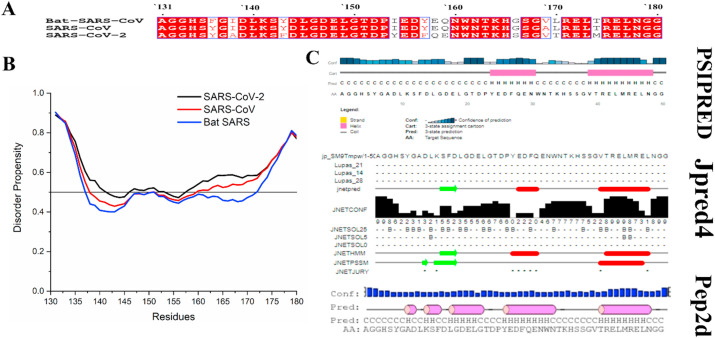
Fig. 2.
(A) The multiple sequence alignment of NSP1-CTR showing consensus sequences. The amino acids conserved in all the analyzed NSP1-CTR sequences are shown in a white with the red background and boxed in black. The residues shown in red with the white background are conserved in most analyzed sequences for their properties. (B) Disorder predisposition predictions of the NSP1-CTR (50 residues) using disorder predictor PONDR® VSL2. The interception at 0.5 scores on Y-axis shows the cut-off of disorderedness. Regions above 0.5 are considered as disordered. (C) Secondary structure predisposition predictions using servers PSIPRED, Jpred4, and Pep2D (C-Coil, E-Extended strand, B-Beta strand, and H-Helix).