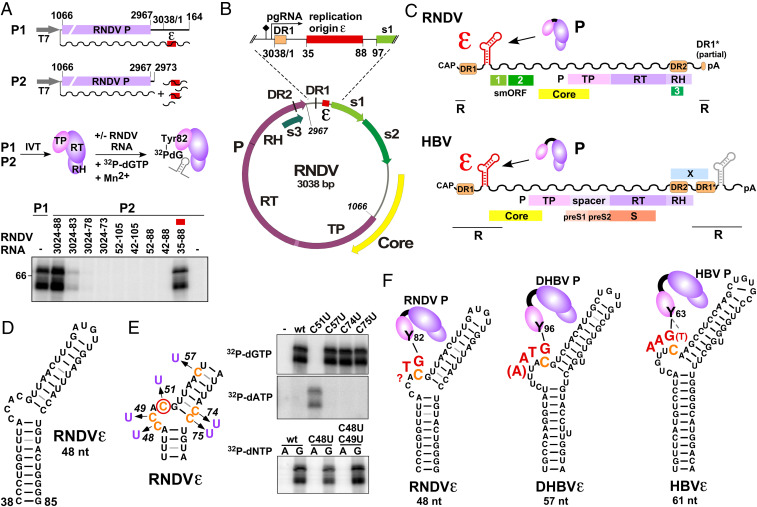
Fig. 1.
Identification and characterization of the RNDV replication origin RNDVε. (A) Mapping of RNDVε (red box). RNDV P was expressed in a coupled in vitro transcription/translation (IVT) system from construct P1 or from P2 complemented with the indicated RNDV UTR RNA fragments (wavy lines, RNA transcripts; T7, T7 promoter). Priming assays were performed, and guanylylation of P was assessed by SDS polyacrylamide gel electrophoresis and phosphoimaging. The top band represents the 72 kDa full-length RNDV P, the band below a truncated yet functional RNDV P translation product. Numbers refer to genomic nt positions as previously defined (2). (B) Position of RNDVε in the RNDV genome. The predicted pgRNA start site and the polyA signal are indicated by arrow and diamond, respectively; s1, s2, and s3 denote nackednavirus specific small ORFs (smORFs) of unknown function. (C) The 5′-proximal position of ε on the pgRNA is conserved between RNDV and HBV. pgRNAs are depicted as wavy lines; terminal redundancy (R), polyA tail (pA). HBV contains a second copy of ε in the 3′ redundancy (gray) dispensible for replication (4). (D) Secondary structure of RNDVε predicted by MC-Fold. (E) The RNDV reverse transcription initiation site maps to RNDVε nt C51. RNDV P was expressed from P2, and nucleotidylation of P was assessed using the indicated C to U point mutants of RNDVε plus [α-32P]dGTP or [α-32P]dATP as substrate. (F) (−)DNA primer (red) and structural context of the primer template of RNDV, DHBV, and HBV. The Tyr residue (Y) of TP, covalently attached to the initiating G residue, is indicated, and the templating C is highlighted in orange. The length of the RNDV primer has not been determined, but the limited complementarity to the putative primer transfer site suggests it does not exceed two nt. HBV possesses a potential alternative initiation site (dashed line to bracketed T).