Extended Data Figure 1. Genomic map of Spjw33 targets and functional outcomes on Ube3a and Snord115 locus.
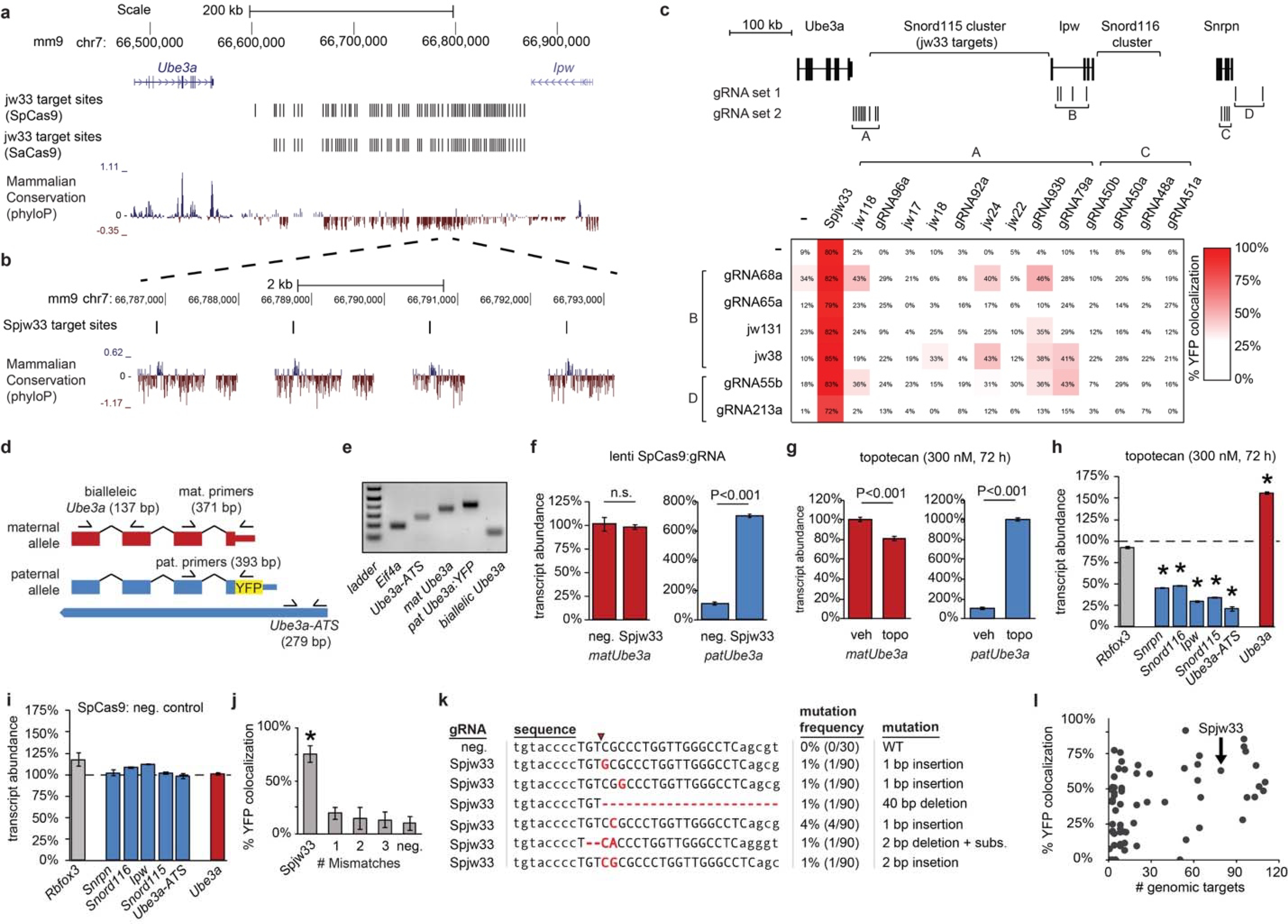
a. Mouse genome browser view.
b. Zoom in showing four Spjw33 target sites. Snord115 genes (blue peaks) are conserved (phyloP track) relative to other regions of Ube3a-ATS.
c. Systematic testing of pairs of gRNAs in Ube3am+/pYFP primary cortical neurons. Top panel: locations of gRNAs positioned to selectively target the Snord115 cluster (gRNA sets A and B), the Snord116 cluster (gRNA sets B and C), or the Ube3a-ATS promoter (gRNA sets A/C and D). Bottom panel: Percentage patYFP+/Cas9+ neurons for each pair of gRNAs relative to the positive control (SpCas9+Spjw33 alone).
d. Location of primers used to quantify transcript expression and discriminate between maternal Ube3a and paternal Ube3a-YFP alleles. Expected band size indicated.
e. Agarose gel showing RT-PCR products amplified from Ube3am+/pYFP cortical neuron cultures. All bands are of the expected size.
f-g. Expression (qPCR) of maternal or paternal Ube3a alleles in cortical neuron cultures transduced with lentivirus carrying SpCas9 and a gRNA targeting human SNORD115 (neg. control), Spjw33 (f), or treated with topotecan (g). Data normalized to Eif4a2, n=3.
h-i. Expression of the indicated genes in wild-type cortical neuron cultures treated with topotecan (h), or lentivirally transduced with SpCas9 and a gRNA targeting human SNORD115 (neg. control) (i). Dashed line marks vehicle (h) or neg. control gRNA (i) expression levels. Normalized to Eif4a2, n=3. * P<0.01.
j. Ube3am+/pYFP neurons transfected with gRNAs containing the indicated number of base mismatches relative to Spjw33. n=4, * P<0.05.
k. Table summarizing mutations identified at the Spjw33 target site. Primary cortical neurons were lentivirally transduced with SpCas9 and either a neg. control gRNA or Spjw33. Genomic DNA spanning the Spjw33 target site was PCR amplified and individual clones were analyzed by Sanger sequencing. Red arrow marks SpCas9 cleavage site. Red letters denote mutations that were introduced by SpCas9, based on the observation that these mutations were not found in neurons treated with the neg. control gRNA nor were they found in other Snord115 genes (mm9 genome build).
l. Percentage YFP colocalization in neurons transfected with SpCas9 and gRNAs targeting the indicated number of genomic sites in the Snord115 locus.
