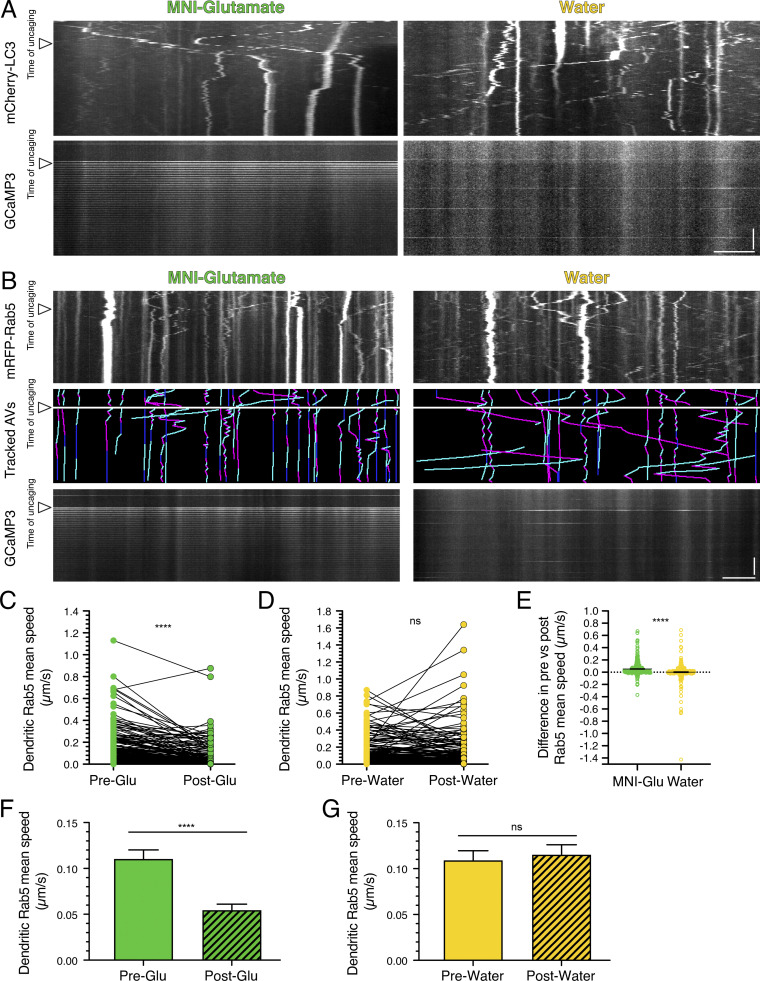
Figure S3.
Local activation of synapses by uncaging MNI-glutamate reduces dendritic AV motility in cortical neurons and early endosome motility in dendrites of hippocampal neurons. (A) Kymograph analysis of mCherry-LC3 and GCaMP3 in dendrites of cortical neurons before and after uncaging (arrowheads denote time of uncaging; water serves as a control). Horizontal bar, 5 µm. Vertical bar, 30 s. (B) Kymographs of mRFP-Rab5, tracked early endosomes (magenta denotes retrograde, cyan denotes anterograde, and dark blue denotes stationary segments), and GCaMP3 from the same dendrite before and after glutamate uncaging (arrowheads denote time of uncaging; water serves as a control). Horizontal bar, 5 µm. Vertical bar, 30 s. (C–G) Quantitation of Rab5 mean speed in dendrites from hippocampal neurons before and during uncaging (C and D, paired points represent a single early endosome before and after uncaging; E, difference in early endosome mean speed before and after uncaging; MNI-glutamate, n = 304 early endosomes from 21 hippocampal neurons; water, n = 313 early endosomes from 20 hippocampal neurons; three independent experiments; 13–14 DIV; C and D, paired t test; E, unpaired t test; ****, P ≤ 0.0001). (F and G) Mean ± SEM of all early endosomes tracked, including those from the paired analysis, before and after uncaging (F, n = 319 early endosomes in pre-uncaging and 340 early endosomes in post-uncaging from 21 neurons from three independent experiments; 13–14 DIV; G, n = 330 early endosomes in pretreatment and 363 early endosomes in post-treatment from 20 neurons from three independent experiments; 13–14 DIV; unpaired t test; ****, P ≤ 0.0001).