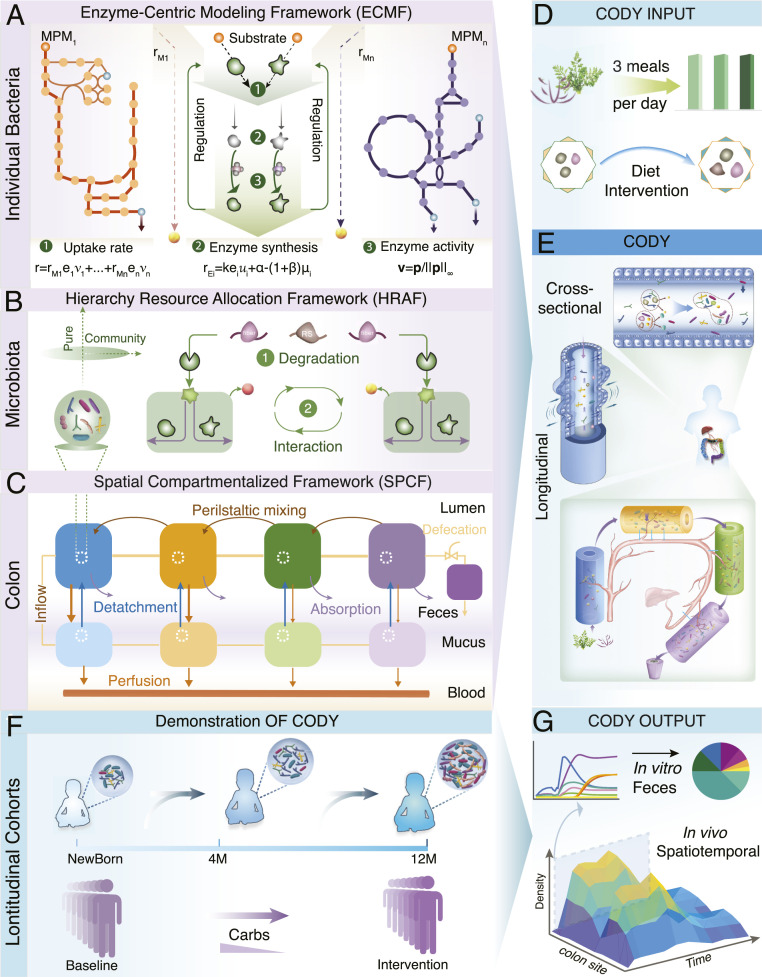
Fig. 1.
Pipeline paradigm of the multiscale framework for COmputing the DYnamics of microbiota (CODY). (A) Schematic of the ECMF for modeling the dynamic growth of individual bacterial (SI Appendix, Supplementary Method 1). Two components of ECMF include metabolism and regulatory processes. A repertoire of MPMs, MPM1 to MPMn, are used to represent bacterial metabolism. These are derived from genome-scale metabolite models. The regulatory process is shown as central green top-down arrows. Kinetic uptake rate of MPMi is shown as by which substrate is converted to products depending on the amount of enzyme assigned to the corresponding MPMi. In the temporal scale, the total uptake rate is distributed among all MPMs in a regulated way (equation 1). Regulation is incorporated by the control variable , which determines enzyme synthesis rate ( in equation 2) and hereby impacts enzyme concentration (), and by the control variable ( in equation 3), which regulates the uptake fluxes (). (B) Illustration of the HRAF for modeling dynamic metabolic behavior of a microbiota community (SI Appendix, Supplementary Method 2). Two interactive levels are shown, i.e., diet-derived microbial-accessible carbohydrate degradation (1) and microbial-microbial interactions (2), with dynamic bacterial biology characterized using ECMF. (C) Schematics of SPCF for biomimetically modeling the colon physiology, including cross-sectional structure with multilayer, water, and metabolite absorption and longitudinal peristalsis movement (SI Appendix, Supplementary Method 3). (D) Daily meals were translated as diet inputs to CODY. (E) Illustration of assembly of the three multiscale frameworks into CODY (SI Appendix, Supplementary Method 8). The nine regions of site-specific residence of the gut microbiota are shown by SPCF, which captures the structure and physiology of the colon. In each region of both longitudinal and cross-sectional directions the intrinsic dynamics of gut microbiota is characterized by microbial–microbial interactions and individual metabolism and impacted by specific colon physiological forces. (F) Evaluation of CODY prediction capacity by demonstrating two longitudinal clinical cohorts involving diet-intervention strategies. The first study involved a long-term gut microbiota dynamic development of Swedish children during early infancy, from neonate to colonization under breastmilk feeding. Afterward, the gut microbiota continued to develop until maturation at 12 mo, when the diet shifted to solid food. The second study involved a short-term gut microbiome dynamic development in response to diet intervention, where obese adults with hepatic steatosis experienced carbohydrate restriction for 14 d. (G) CODY enables predictions on the spatiotemporal dynamic variations of in vivo gut microbiota, in vitro microbiota composition in the feces, and associated metabolite changes.