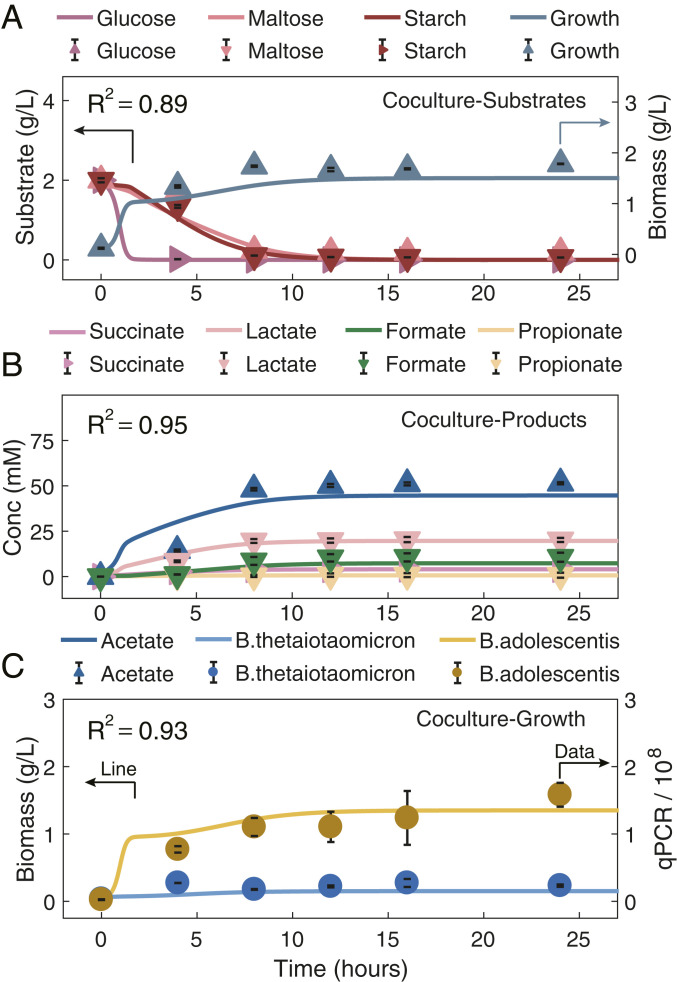
Fig. 2.
Evaluation of HRAF prediction capacity in characterizing dynamic microbial–microbial interactions and growth dynamics of the synthetic microbial coculture, through comparing pure model predictions on dynamic nutrient utilization profiles, dynamic growth profiles, and microbial–microbial interaction profiles of the two-species coculture with experimental data. Evaluation of HRAF’s prediction capacity in characterizing the dynamic nutrient profiles (A), dynamic growth profiles (B), and individual microbial growth capacity of a two-species cocultures, i.e., Bth and Bad, with the use of an experimental dataset (C). In each panel, the HRAF prediction results are shown as solid lines, and experimental data are shown as symbols. Different microbial-associated metabolite and growth profiles are denoted by specific colors presented above each panel. A shows the comparison results between the HRAF predictions on the dynamic growth profiles and utilization profiles of three substrates, glucose, maltose, and starch, and the experimental dataset for the two-species cocultures. B indicates the comparison results between HRAF predictions and experimental dataset on associated dynamic metabolite profiles for the two-species cocultures. C exhibits the comparison results between HRAF predictions and experimental data on individual microbial growth profiles of the two-species coculture. The comparison results between model predictions and experimental data are evaluated and measured by the coefficient of determination (R2; the calculation equation can be found in Methods) in each panel ranging from 0.89 to 0.95.