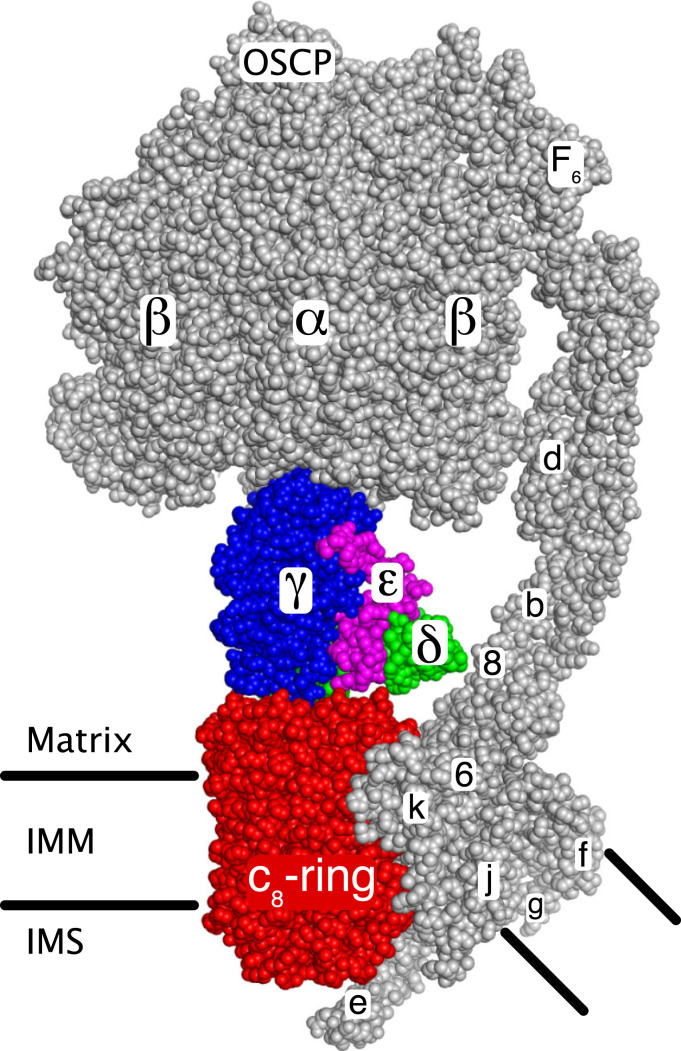
Fig. 1.
The rotor and stator components of monomeric bovine ATP synthase. The two main functional components of the enzyme, the rotor (colored) and the stator (gray), are shown. The rotor consists of the c8-ring (red) associated with the IMM, and the central stalk consisting of subunits γ (blue), δ (green), and ε (magenta). The upper region of the γ-subunit (view obscured) is asymmetrical and penetrates into the upper spherical part of the stator, consisting of three α-subunits and three β-subunits where the three catalytic sites of the enzyme are found. The spherical region and the central stalk together are known as the F1-domain. The F1-domain and the upper part of the associated PS on the right, extend into the mitochondrial matrix. The PS, made of the oligomycin sensitivity conferral protein (OSCP) and subunits b, d, and F6, is attached via the OSCP to the upper surface of the spherical region and extends into the IMM. The membrane domain of the PS is associated with subunits e, f, g, j (or 6.8-kDa proteolipid), ATP6 (or 6), and ATP8 (or 8). The membrane region consisting of subunits e, f, g and the transmembrane α-helices of subunits b and associated lipids is referred to as “the wedge” (23). Subunit k (or diabetes-associated protein in insulin sensitive tissue, DAPIT) is bound to ATP6, and subunit e extends into the intermembrane space (IMS). The turning of the rotor, driven by energy from the transmembrane proton motive force, brings about a series of structural changes in each of the three catalytic sites leading to the binding of substrates and the formation and release of three ATP molecules for each 360° rotary cycle. Image credit: M. G. Montgomery (University of Cambridge, United Kingdom).