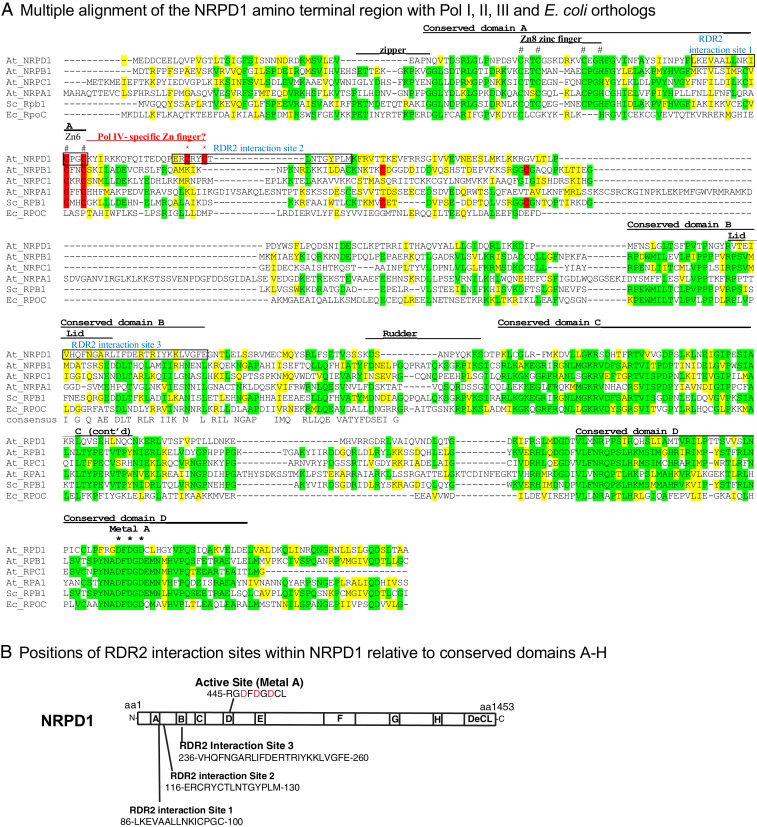
Fig. 7.
Comparison of amino terminal regions of RNA polymerase largest subunits. (A) Multiple alignment of NRPD1’s amino terminus with corresponding sequences of the largest subunits of Arabidopsis (At) Pols I (NRPA1), II (NRPB1), and III (NRPC1); budding yeast Pol II (Sc_RPB1); and E. coli polymerase (Ec_RpoC). Identical amino acids in three or more sequences are shaded green; similar amino acids are shaded yellow. Positions of conserved domains A–D, zipper, lid, and rudder elements are indicated based on their positions in yeast Pol II. The position of the Metal A site is also indicated, with black asterisks denoting the three magnesium-binding aspartates. Conserved Zinc coordinating amino acids of Zn8 and Zn6 are indicated with hash symbols (#). Cysteines of Zn6 are further highlighted in red, with cysteines of the Pol IV-specific CxxC, motif downstream of the conserved CxxC motif, indicated with red asterisks. RDR2 interacting regions 1, 2, and 3 of NRPD1, which overlap both Zn6 CxxC motifs and the lid element, respectively, are indicated in blue. (B) Relative locations of the active site and RDR2-interacting regions in a linear depiction of full-length NRPD1. Positions of domains A–H, conserved among largest subunits of all multisubunit RNA polymerases, and the C-terminal DeCL domain (also present in the Pol V largest subunit) are highlighted.