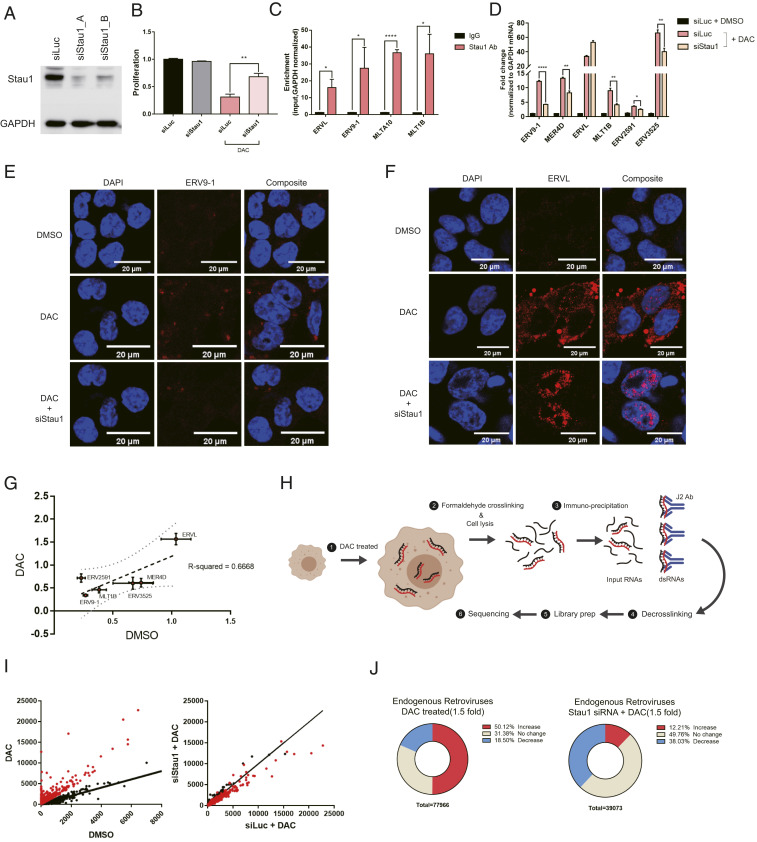
Fig. 2.
Stau1 interacts with ERVs and regulates their expression and localization. (A) Western blot result to confirm the knockdown efficiency of siRNAs used in this study. siStau1_A targets the 3′ UTR of Stau1 mRNA while siStau1_B targets the coding region of the mRNA. We present the data using siStau1_A for the rest of the study, but siStau1_B also yielded similar results. (B) The knockdown of Stau1 significantly rescued cell death from decitabine treatment. n = 3 and error bars denote SEM. (C) Direct interaction between Stau1 and ERV RNAs was confirmed using RNA immunoprecipitation (RNA-IP) after formaldehyde cross-linking. n = 3 and error bars denote SEM. Enrichment relative to that of GAPDH mRNA is shown. (D) Knockdown of Stau1 resulted in decreased expression of most ERV RNAs examined. n = 3 and error bars denote SEM. (E and F) RNA FISH of ERV9-1 (E) and ERVL (F) revealed that Stau1 could affect the expression and subcellular localization of ERV RNAs. (G) Stau1 knockdown also affected the basal expression of ERV RNAs without decitabine. Fractional change of ERV RNA expression levels assessed by RT-qPCR for DMSO control and decitabine-treated samples is shown. The two conditions show a good positive correlation with R2 = 0.6668. (H) A schematic for the J2 fCLIP dsRNA-seq process. (Created with BioRender.com.) (I) Distribution of ERV RNA expression in the J2 dsRNA-seq result. Each dot represents sequencing reads mapped to a specific ERV locus. Red dots represent ERV loci with at least a 1.5-fold increase in expression by decitabine. (J) Pie chart of percent of ERV loci with a change in expression by decitabine and in Stau1 knockdown cells. *P value <0.05, **P value <0.01, ***P value <0.001, ****P value <0.0001.