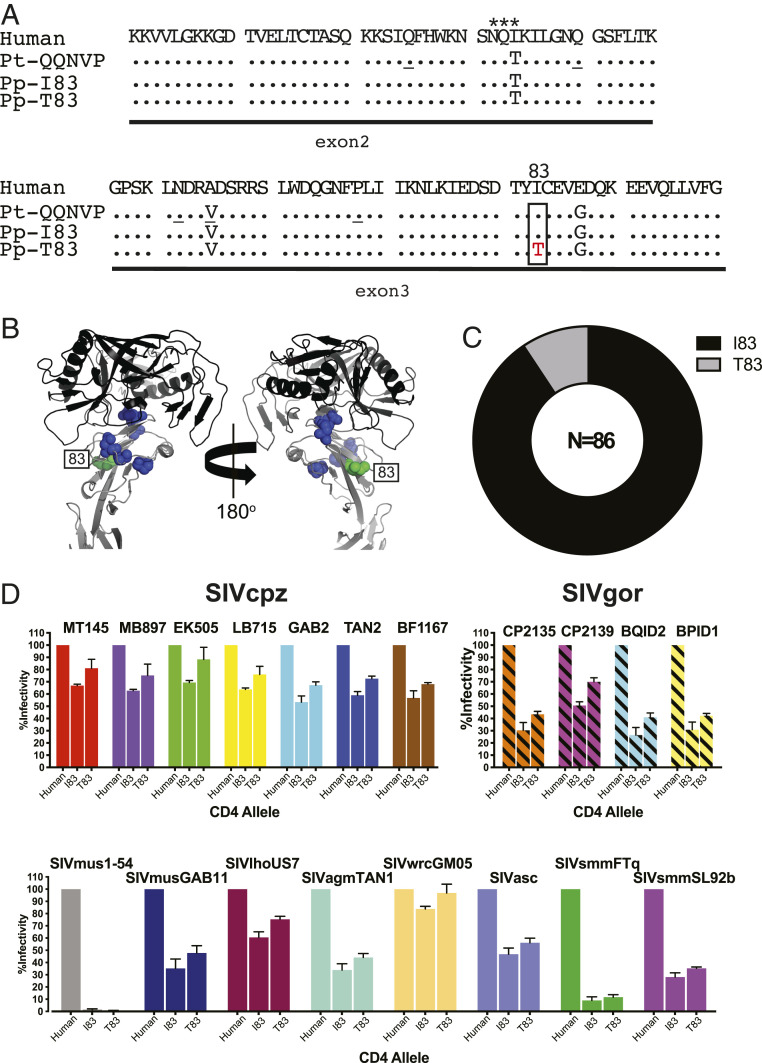
Fig. 1.
Allelic diversity of CD4 in bonobos. (A) CD4 coding variants identified in wild bonobo populations. D1 domain variants of bonobos (P. paniscus, Pp; derived from exon 2 and 3 sequences) are compared to human and chimpanzee CD4, with dots indicating identity to the human reference. A single amino acid replacement (I83T) in the bonobo CD4 is boxed. A potential N-linked glycosylation site is indicated by asterisks. The ancestral chimpanzee (P. troglodytes; Pt) allele is shown for reference, with positions that are polymorphic in chimpanzees underlined. (B) Crystal structure of the HIV-1 gp120 envelope domain (black) bound to human CD4 (gray) (PDB ID code 4R2G), with polymorphic positions indicated for chimpanzees (blue) and bonobos (green). (C) CD4 allele frequency in wild bonobos. The number of tested individuals is indicated. (D) Effects of bonobo CD4 polymorphisms on SIV Env mediated cell entry. The infectivity of pseudoviruses carrying different SIV Envs is shown for transiently transfected cells expressing human and bonobo CD4 variants and the cognate CC5 receptors. Values are scaled relative to the human CD4 (set to 100%). Bars represent the average of three independent transfections, each performed in triplicate, with SDs shown (fold-changes of Env infectivity for different CD4 alleles are shown in SI Appendix, Table S2).