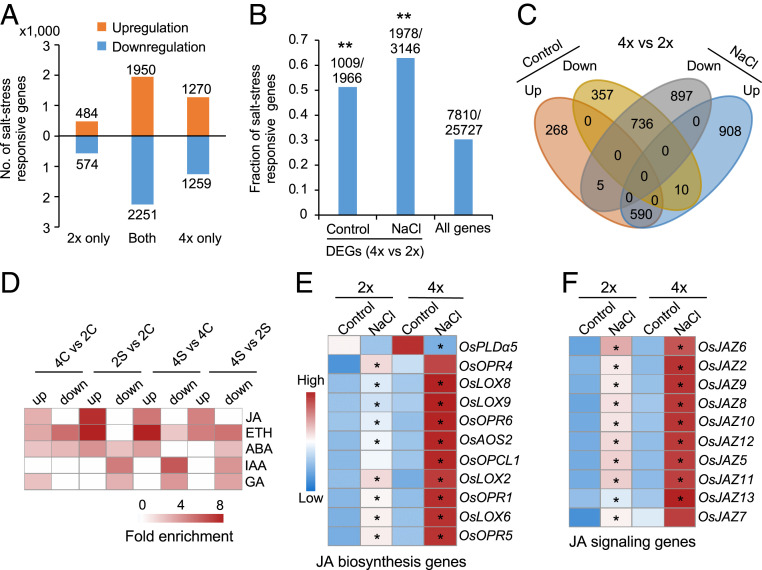
Fig. 2.
Polyploidy primes activation of JA-related genes under salt stress in 02428. (A) Number of DEGs in diploid and tetraploid rice under salt stress, which were divided into up- or down-regulated DEGs in diploid (2×) only, tetraploid (4×) only, or shared (both). (B) Ploidy-dependent genes are associated with salt stress–responsive genes. Y-axis shows fraction of salt responsive genes, with the numbers above columns indicating salt responsive genes over DEGs between tetraploid (4×) and diploid (2×) rice in the control or salt (NaCl) condition or all expressed genes. Double asterisks indicate the significance level of P < 0.01 (hypergeometric test). (C) Venn diagram showing overlap between ploidy-dependent genes without (control) or with (NaCl) salt stress, which are concerted up- or down-regulated in response to salt treatment. (D) Enrichment of phytohormone-related genes in ploidy-dependent and salt responsive genes in tetraploid or diploid rice without salt (control, 4C versus 2C) or with salt (4S versus 2S) stress. JA: jasmonic acid; ETH: ethylene; ABA: abscisic acid; IAA: indole-3-acetic acid; GA: gibberellic acid. (E and F) Expression levels of JA biosynthesis (E) and signaling (F) related genes in diploid (2×) and tetraploid (4×) rice without (control) or with salt (NaCl) treatment. A single asterisk indicates gene expression level difference before and after salt treatment in diploid and tetraploid rice, respectively (FDR < 0.05 and log2[fold change] ≥ 1).