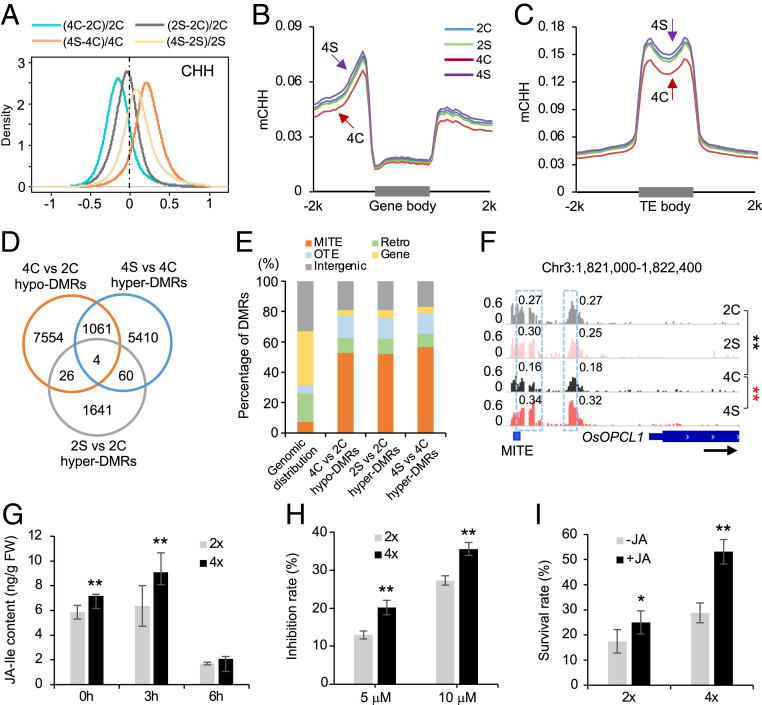
Fig. 3.
Dural roles of CHH methylation in activation of JA-related genes and repression of TEs in 02428. (A) Global CHH methylation changes after polyploidization and salt stress. Diploid rice is without (2C) or with salt (2S) treatment, and tetraploid rice is without (4C) or with salt (4S) stress. (B and C) Distributions of CHH methylation in protein-coding genes (B) and TEs (C). (D) Venn diagram showing overlap among ploidy-induced hypo-DMRs and salt-induced hyper-DMRs. (E) Distribution of stress-related hypo- or hyper-DMRs in miniature inverted transposable element (MITE), retrotransposon (Retro), other TE (OTE), gene, and intergenic regions, relative to their genomic distributions. (F) An example (genome browser view) of CHH methylation changes in the promoter of OsOPCL1. The gene and a MITE element are shown with a black arrow indicating transcriptional direction. Numbers in snapshot indicate CHH methylation levels of regions in dotted box. Double asterisks indicate the difference between 4C and 2C (black) and between 4S and 4C (red), respectively, at a statistical significance level of P < 0.01 (Fisher’s exact tests). (G) JA-Ile content in diploid and tetraploid rice under saline condition for 0, 3, and 6 h (n = 3 biological replicates). (H) Inhibition rates of diploid and tetraploid rice after treatments with 5 or 10 μM JA for 2 d (n = 30 biological replicates). (I) Survival rates of diploid and tetraploid rice after salt stress are increased by the treatment of 0 or 1 μM JA (n = 5 replicates, each replicate with 80 plants). Single and double asterisks indicate statistical significance of P < 0.05 and P < 0.01, respectively (Student’s t test).