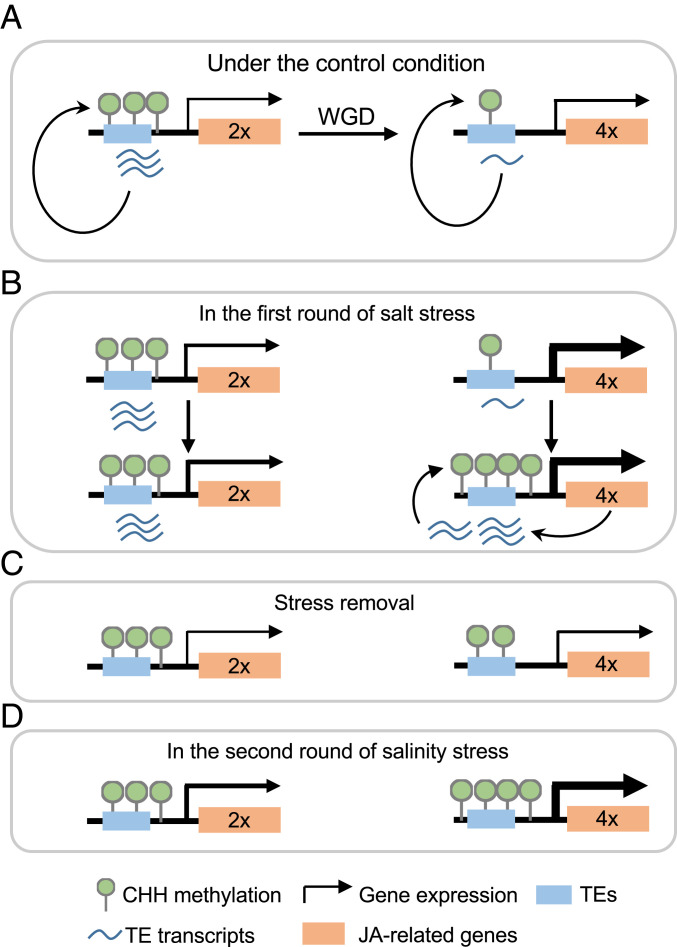
Fig. 6.
Models for enhanced salt tolerance and fitness in tetraploid rice. (A) Polyploidy may repress proximal TEs through histone modifications, leading to CHH hypomethylation by RdDM pathway in tetraploid rice. (B) Under salt stress (the first salt treatment), CHH hypomethylation primes stress-related genes including those in JA biosynthesis and signaling pathways for more rapid and stronger activation of these genes to enhance salt tolerance in tetraploid (Upper) than in diploid rice. In turn, highly expressed stress-related genes may derepress neighboring TEs, triggering CHH hypermethylation to silence these TEs (Lower). (C) After stress removal, transcript abundance of JA-related genes in tetraploid returns to a similar level to diploid rice, while CHH hypomethylated status of tetraploid rice prior to the first salt treatment are also partly retained. (D) After recurring salt stress, stress-related genes of tetraploid rice are more strongly induced and subsequently hypermethylated in a process similar to that in B.