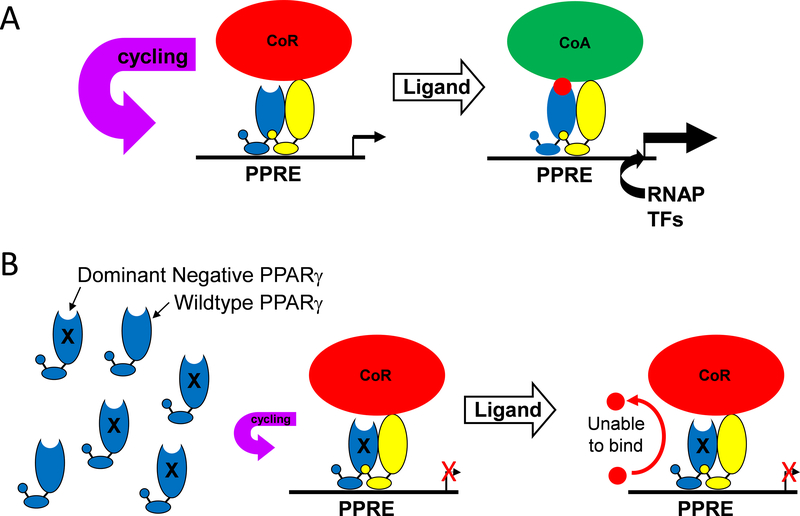
Figure 2: Mechanism of Dominant Negative PPARγ Mutations.
A) As illustrated in Figure 1, PPARγ forms a heterodimer with RXR and binds to a PPAR response element (PPRE) in chromatin in the regulatory region of a PPARγ target gene even in the unliganded state. This is not a static process, because the complex cycles through DNA bound and unbound states. Consequently, the rate of transcription under these circumstances reflects the occupancy of the site by the PPARγ-RXR-corepressor complex and the binding of other stimulatory transcription factors and RNA polymerase to other sequences in the regulatory region of the gene. Ligand binding causes dismissal of the corepressors (CoR) and a recruitment of coactivators (CoA). The coactivator complex facilitates a conformation of chromatin favorable for transcription. B) Replacement of wild-type PPARγ with P467L PPARγ causes increased occupancy (reduced cycling) of the PPRE by the PPARγ-RXR-corepressor complex, which further reduces baseline transcription of actively repressed PPARγ target genes. Figure was previously reported in 46.