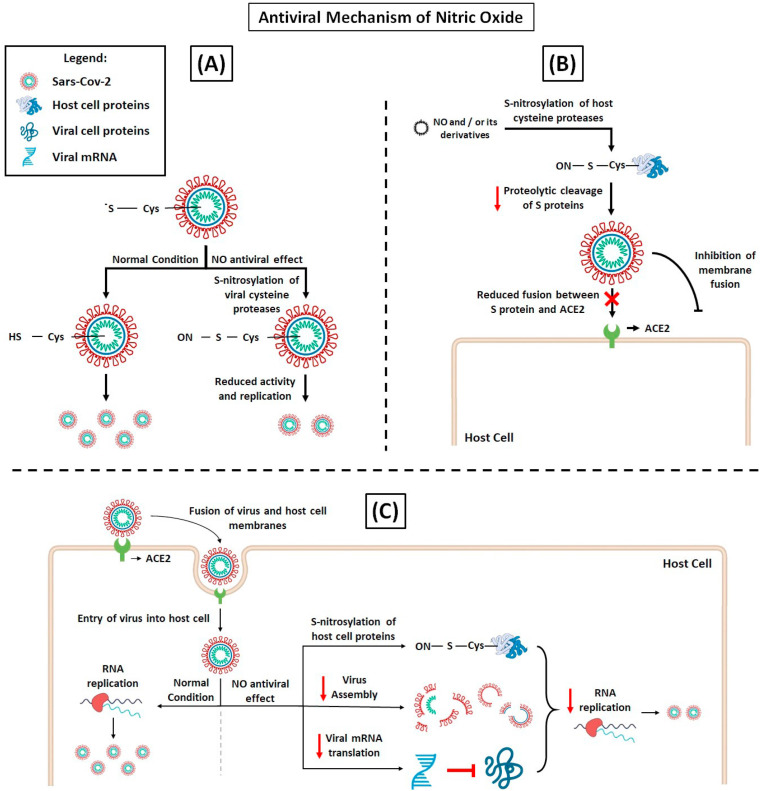
Fig. 2.
NO antiviral mechanisms, hypothesis of action on SARS-CoV-2 replication. (A) Acting on viral proteases. The processing of the polyprotein region is a point of posttranslational control that is essential for virus replication. SARS-CoV-2 processes the polyproteins using two cysteine proteases, the papain-like protease (PLpro) or the chymotrypsin-like protease (Mpro). S-nitrosylation of specific Cys residue reduces the activity of these proteases inhibiting SARS-CoV-2 replication [106,107]. (B) Acting on host cell proteins. The complete intracellular life cycle of SARS-CoV-2 relies on interactions with host molecules. The proteolytic cleavage of S proteins by serine protease TMPRSS2 and cysteine proteases cathepsin B (CatB) and CatL is essential for the virus fusion. Thus, the inhibition CatB and CatL by S-nitrosylation could prevent SARS-CoV-2 entry into cell. (C) Furthermore, NO-mediated S-nitrosylation of cysteine-containing proteins may prevent virus molecular interactions critical for RNA replication, virus assembly and translation of viral mRNAs, abrogating SARS-CoV-2 cell cycle [108,109].