Figure 2.

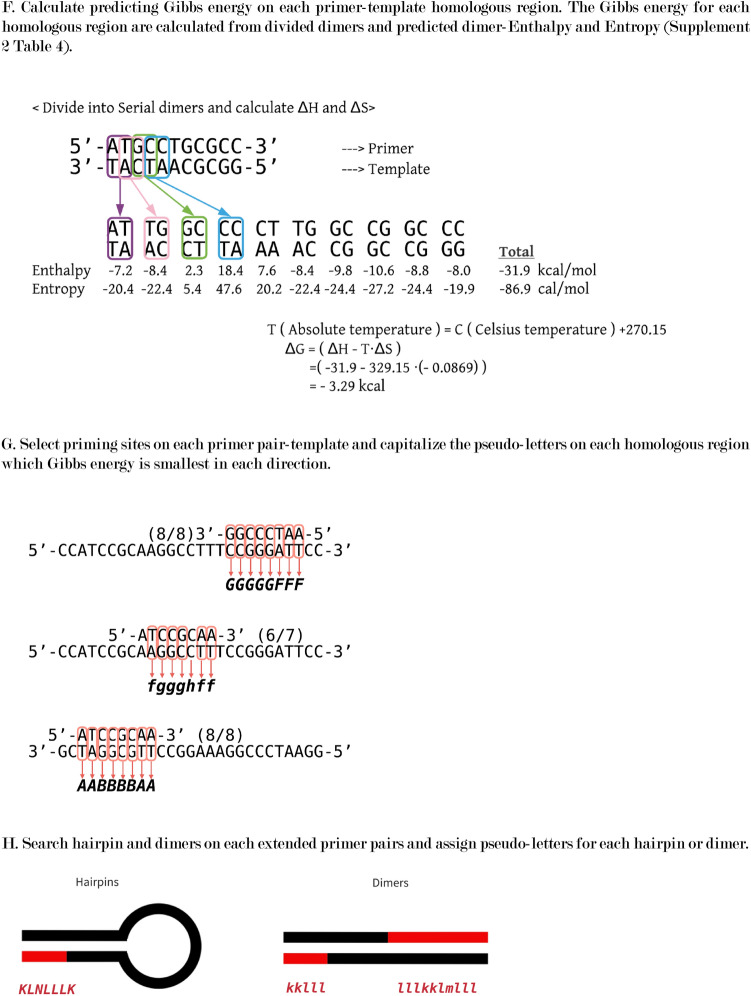
The process of generating pseudo-words and pseudo-sentences is shown. Pseudowords are generated in relation to a particular primer pair and template. First, prepare the primer pair and template data in a format that can be read by the analysis program (A). Then, the base sequence alternatives which synthesized on the primer hairpin (B) and dimer (C) are added to the original primer sequences. The plausible double-strand formation which is expected between the primer sets and template is assumed and expressed as letters (D–E). First, a part of the complementary primer including a part of the primer and the template and the position of the template are listed (D), and their interaction is expressed by a letter for each base-pair (E). The one-character code used to express the interaction used at that time is shown in E. In order to do machine learning with RNN, it is necessary to predict the primer-binding position on the template, which is the source of the PCR product production. On the prediction other primer-binding positions are classified to unrelated binding positions the PCR product production. In this study, the free energy of each plausible primer binding position on the template was calculated for all possible primer binding positions. Referring to the free energy of binding positions, two primer binding positions, which have minimum free energy, were identified as the PCR-amplifiable primer binding positions. For these determinations, the free energy was calculated on nested dimers and sum free energies on the primer-template binding positions (F). The free energies are calculated from Enthalpy, Entry, and absolute temperature of the nested dimers. According to the free energies on the primer-template binding positions, we determined two primer-template binding sites, from which PCR is most likely to proceed, and capitalize nucleotide-interaction-letters (G). Similar to primer-template interactions, the program searches hairpin or dimer formation in a primer and primers. One-letter codes are generated for each base pair in these hairpin and dimer (H). Strings of interactions between primers or between primers and templates were broken down into 5 letters (five-character codes) as words and duplicated to reflect their importance depending on their length and position from the 3'end (I). Similarly, the interaction is predicted for the PCR product and primers shown in Fig. 1D, and characters different from the interaction assumed in the middle of the process are assigned (J). A pseudo-sentence is generated by arranging all the five-character codes assigned in this way at positions based on the array of templates (K).