Fig. 7. Models of monomeric and dimeric DNA-PKcs showing the positions of the YRPD motif and the ABCDE loop.
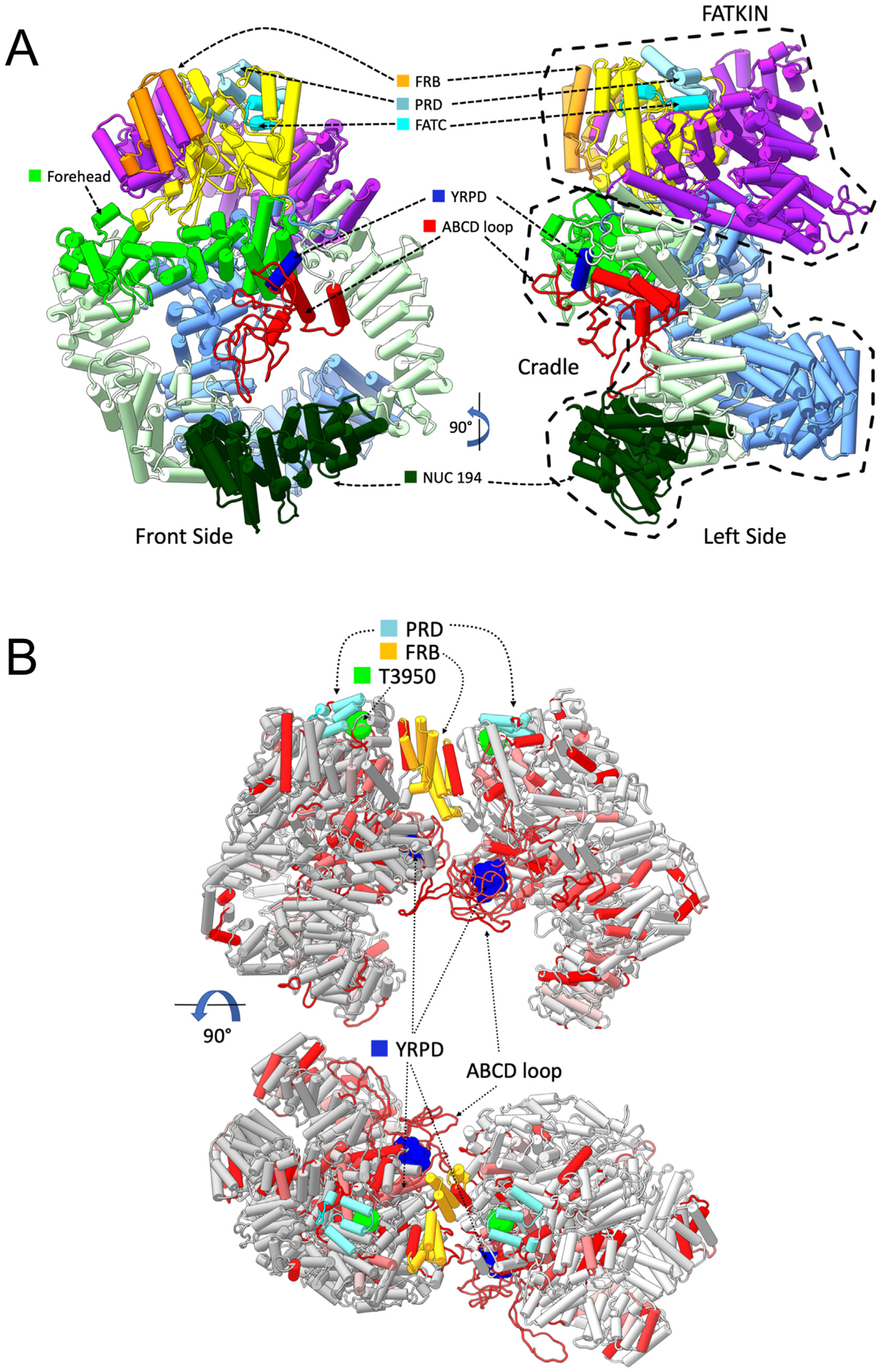
Panel A: DNA-PKcs model with added missing regions shown in pipes and planks representation. The model of DNA-PKcs was built based on the X-ray structure (PDB:5LUQ from (Sibanda et al., 2017)) by adding missing loops using MODELLER (Sali and Blundell, 1993), including the ABCDE loop region (red). Major domain in DNA-PKcs are coloured as highlighted, where the N-HEAT domain is shown in blue, the M-HEAT in green, the FAT in purple, the kinase in yellow and the FATC in bright light blue. Also shown is the location of the NUC194 domain (dark green) and the Forehead domain (light, bright green). The added disordered loop containing the ABCDE sites is shown in red. The conserved YRxGxxPD sequence beginning at Y2776 is shown in dark blue. Also shown are the FRB (orange) and PRD (grey/blue) motifs.
Panel B: The locations of highly conserved regions from selected metazoa were mapped onto the DNA-PKcs dimer model (see Methods and Hammel et al., submitted, this issue). Amino acids that were identical and/or highly conserved (>95%) in all vertebrate/invertebrates examined (Supplementary Fig. 2) were mapped onto two orthogonal views of dimer models shown in pipes and planks representation. Regions containing identical or conserved regions are coloured in red and light red, respectively. The highly conserved YRPD residues (dark blue) are shown in sphere representation. Phosphorylation site threonine 3950 is in bright green, the FRB in orange and the PRD in grey/blue. The position of the ABCDE loop is shown in red.
