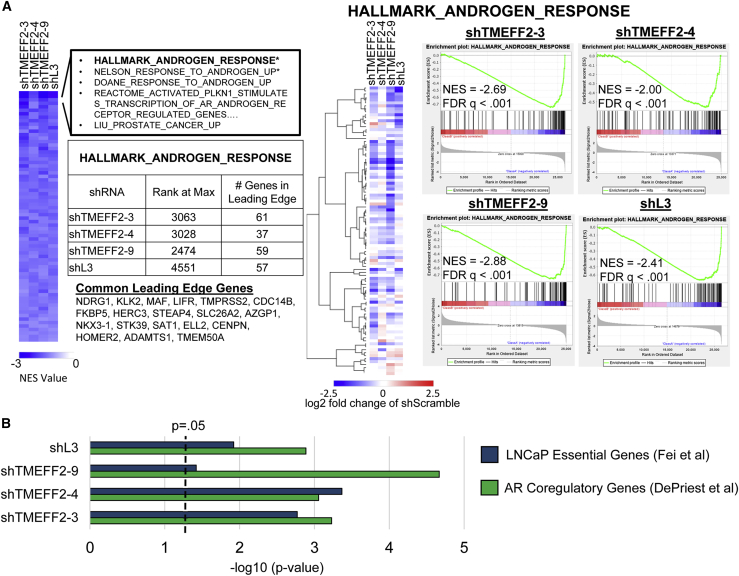
Figure 3.
TMEFF2-targeted shRNAs and shL3 inhibit androgen transcriptional response and downregulate AR coregulatory and essential genes
(A) Heatmap (left panel) shows normalized enrichment scores (NES) of common significantly enriched gene sets (q value < 0.25, for each shRNA) from gene set enrichment analyses (GSEAs) of RNA-seq data from LNCaP cells expressing designated shRNAs, compared to cells expressing the shScramble control. Negative NES indicates gene sets are enriched among downregulated genes relative to shScramble control cells. Heatmap (middle panel) shows relative gene expression of genes within the hallmark androgen response gene set. GSEA enrichment plots (right panel) show enrichment of the Hallmark Androgen Response gene set within gene lists rank ordered by gene expression (upregulated to downregulated) in cells expressing designated shRNAs relative to shScramble control cells. NES and p value are labeled on each plot. The number of Hallmark Androgen Response genes within the leading edge for each shRNA is indicated in the table, and the 19 genes located in the leading edge for all shRNAs are listed. (B) –log10 p value for enrichment of AR coregulatory genes19 and LNCaP essential genes20 among significantly downregulated genes by each shRNA, relative to shScramble expressing LNCaP cells. –log10 p values are based on hypergeometric distribution.