Figure 4.
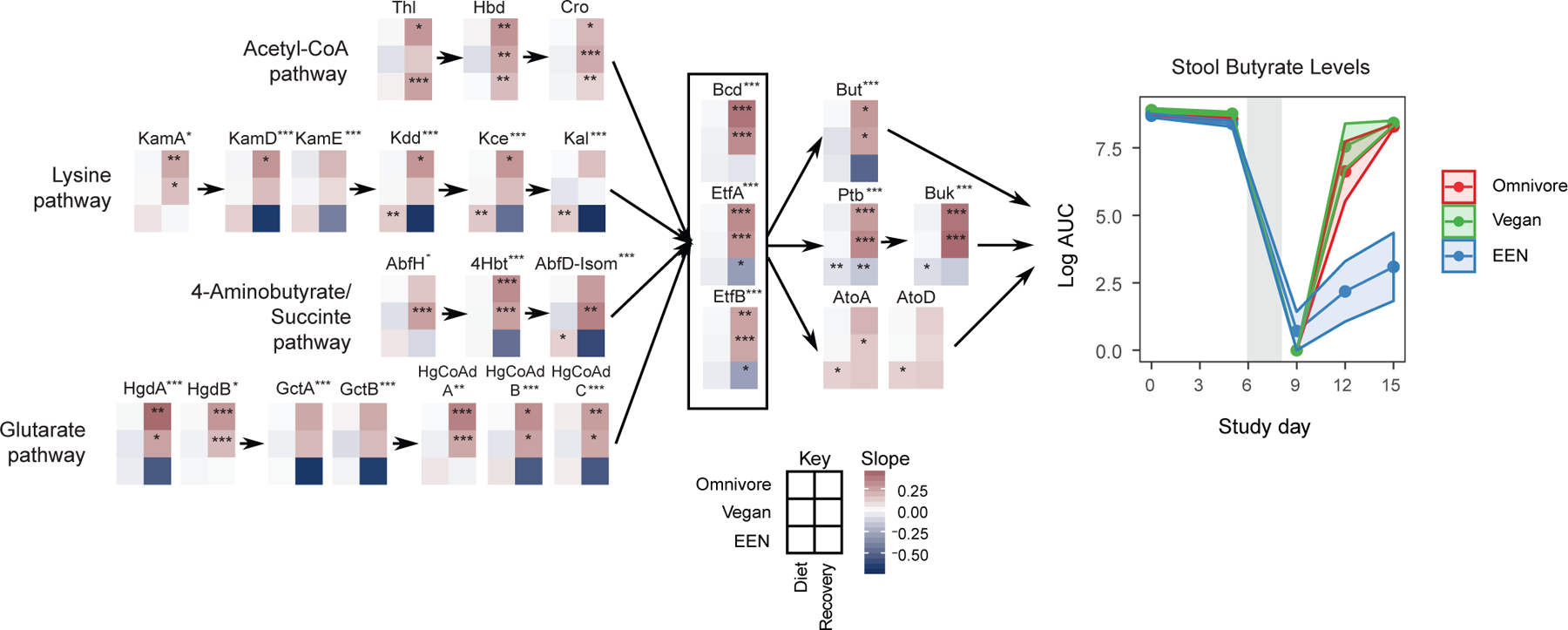
The human gut microbiome and butyrate production. Left Panel: The change in relative abundance of genes in four pathways responsible for butyrate production as curated by Vital et al.(Vital et al., 2014). Linear mixed effects models were fit to logit transformed gene relative abundance levels for each diet and study phase (diet or recovery) separately. Each 6 grid heatmap represents the slopes obtained from these linear mixed effects models where the rows represent the diet slopes and the columns represent the study phase. The stars within the heatmap boxes represent if the slope is significantly different than 0 based on linear mixed effects models. The p values were corrected for false discovery rate using Benjamini-Hochberg method (q<0.05). A separate linear mixed effects model was built to find the genes that show a different slope profile in EEN group compared to the omnivore group during the recovery phase. The stars next to the gene names represent if this progression profile of gene abundance is significantly different (*q<0.05, **q<0.01, ***q<0.001). Right Panel: Fecal butyrate levels throughout the duration of the FARMM study from untargeted metabolomics results. The confidence intervals represent the SEM. The gray shaded areas represent the antibiotic/PEG phase of the study.
