Figure 3.
Generating chromosome-free cells that remain metabolically active
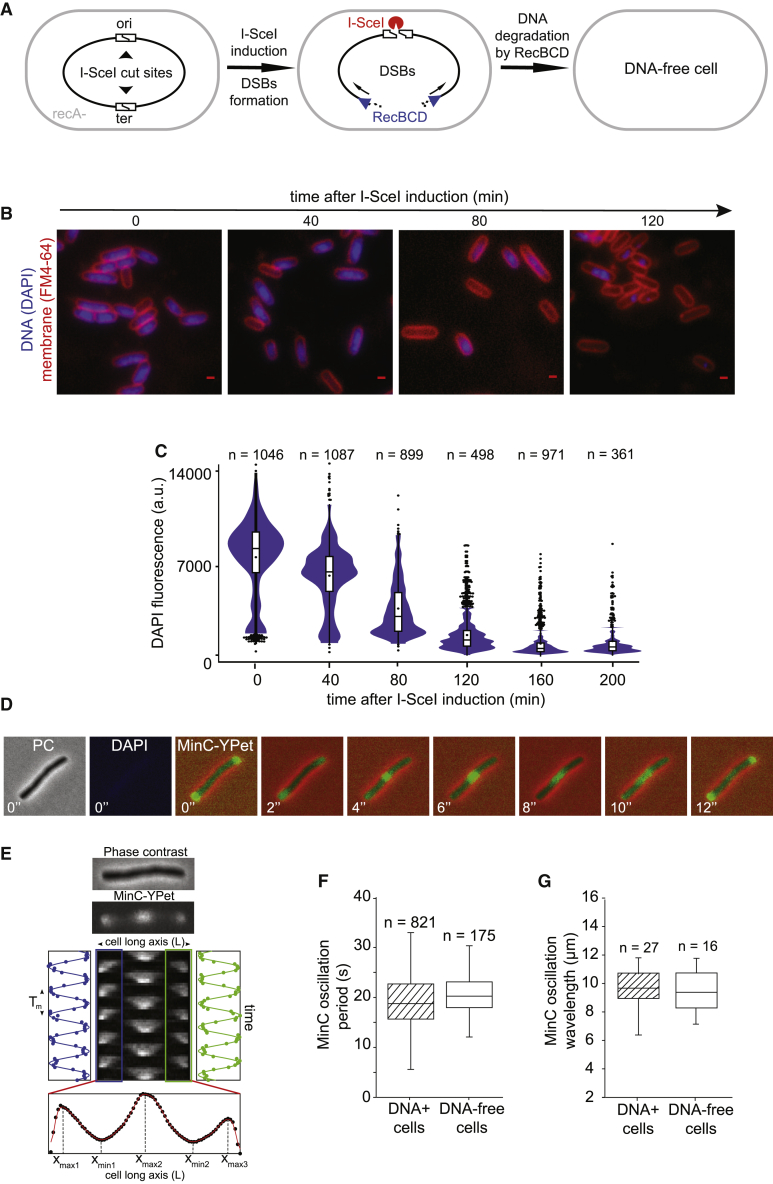
(A) Schematic of the chromosome-degradation system. Induction of I-SceI endonuclease causes 2 double-stranded-breaks (DSBs) in I-SceI cut sites at diametrically opposed positions on the chromosome. In a recA strain, processing of DSBs by RecBCD results in complete chromosome degradation.
(B) Chromosome degradation after I-SceI induction, revealed by loss of DAPI-stained DNA fluorescence (blue) in cells with an FM464-labeled membrane (red). Scale bar, 1 μm.
(C) DAPI fluorescence profiles show complete chromosome degradation 120 min after I-SceI induction (black dot, mean and outliers; horizontal lines, median, first and third quartiles; n, number of cells analyzed).
(D) MinC-YPet oscillation in an example chromosome-free cell. Cell filamentation was induced by cephalexin treatment. Transmitted light, DAPI, and MinC-YPet fluorescence images were obtained 120 min after I-SceI induction. Scale bar, 1 μm.
(E) Kymograph of MinC-YPet oscillation in an example filamentous cell. Kymograph width corresponds to the long cell axis (L). Time-dependent intensity in the cell halves (blue, green) shows the oscillation period Tm. The time-average profile underneath shows the oscillation wavelength.
(F and G) MinC-YPet oscillation period and wavelength are similar with and without chromosome degradation (n, number of cells analyzed, error bars: standard deviation [STD]).
See also Figures S2–S5.