Figure 6.
Quantitative partitioning of protein states
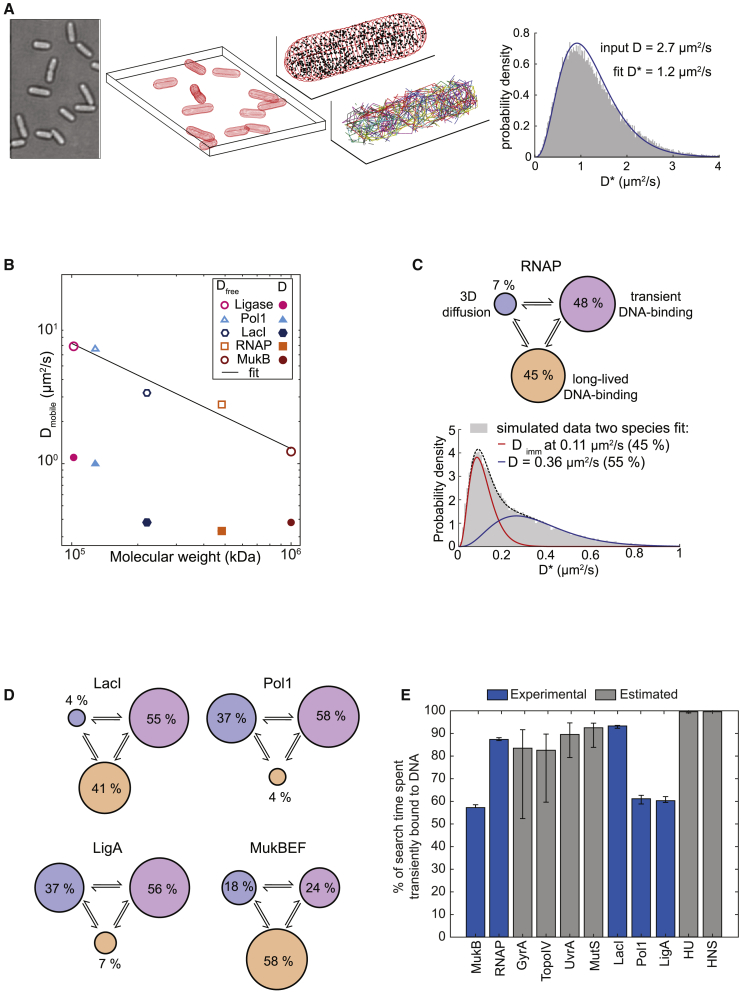
(A) Illustration of Brownian motion simulation to estimate the unbiased diffusion coefficients Dmobile (in unperturbed cells) and Dfree (in chromosome-free cells).
(B) Dmobile and Dfree plotted versus molecular weight M on a log scale. Linear fit log(Dfree) = α∙log(c∙M).
(C and D) Partitioning long-lived DNA-binding (orange), transient DNA-binding (purple), and 3D diffusion (blue) states for RNAP and (D) for LacI, Pol1, LigA, and MukB.
(E) The percentage of search time spent bound non-specifically to DNA for all 11 studied DNA-binding proteins. Blue bars show the proteins with Dfree measured in chromosome-free cells, and gray bars show proteins with Dfree estimated from the fit in (B). Error bars: STD.
See also Figure S6.