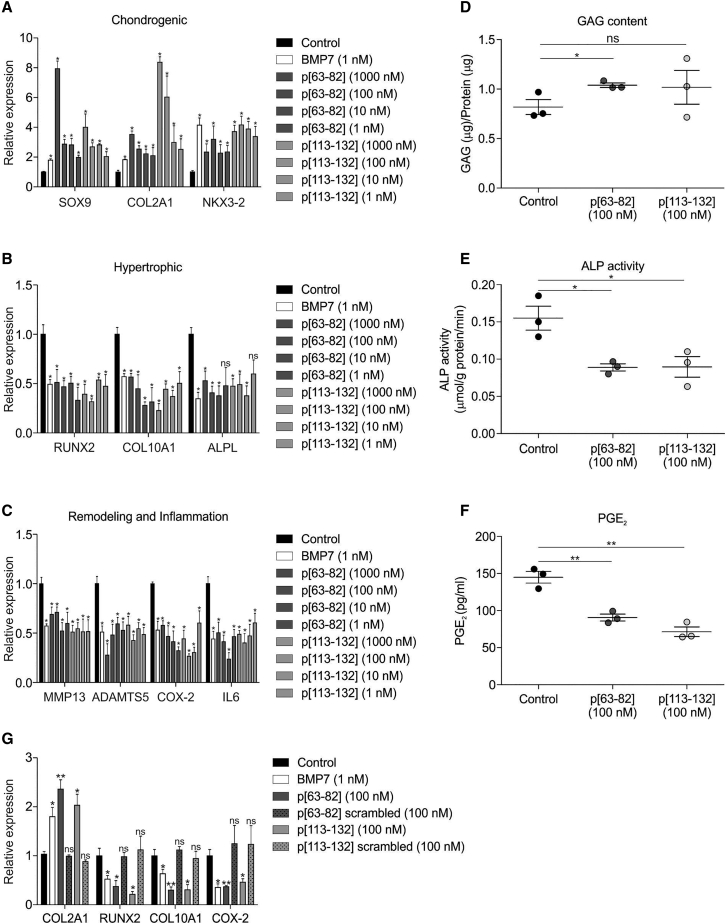
Figure 2.
Bioactivity of BMP7-derived peptides p[63−82] and p[113−132] on OA-HACs
Complete peptide library screening quantitative real-time PCR dataset for peptides p[63−82] and p[113−132] is presented. (A) SOX9, COL2A1, and NKX3-2 mRNA expression in the 18 OA-HAC pool exposed to 1,000, 100, 10, or 1 nM of peptide p[63−82] and p[113−132] is shown and presented relative to control (black bars) and alongside full-length BMP7 (white bars). (B) Similar to (A) but for chondrocyte hypertrophy-associated genes RUNX2, COL10A1, and ALPL. (C) Similar to (A) but for cartilage extracellular matrix remodeling-associated genes MMP13 and ADAMTS5 and inflammation-related genes COX-2 and IL-6. (D) Glycosaminoglycan (GAG) content (normalized to total protein content) on an independent cohort of three individual OA-HAC donors treated with 100 nM peptides p[63−82] and p[113−132] for 24 h. Corresponding gene expression data are shown in Figure S3. (E) Alkaline phosphatase (ALP) activity (normalized to total protein content) in similar samples from (D). (F) Prostaglandin E2 (PGE2) levels in culture supernatant in similar samples from (D). (G) In the same OA-HAC pool of 18 OA-HAC donors from the peptide library screening, sequence dependency of peptides p[63−82] and p[113−132] was determined by exposing the OA-HAC pool to scrambled versions of peptides p[63−82] and p[113−132] (100 nM, 24 h). Expression of COL2A1, RUNX2, COL10A1, and COX-2 mRNAs is shown. Error bars represent mean ± SEM, and statistical significance for peptide conditions or BMP7 versus control condition as determined by unpaired two-tailed Student’s t test is represented as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.