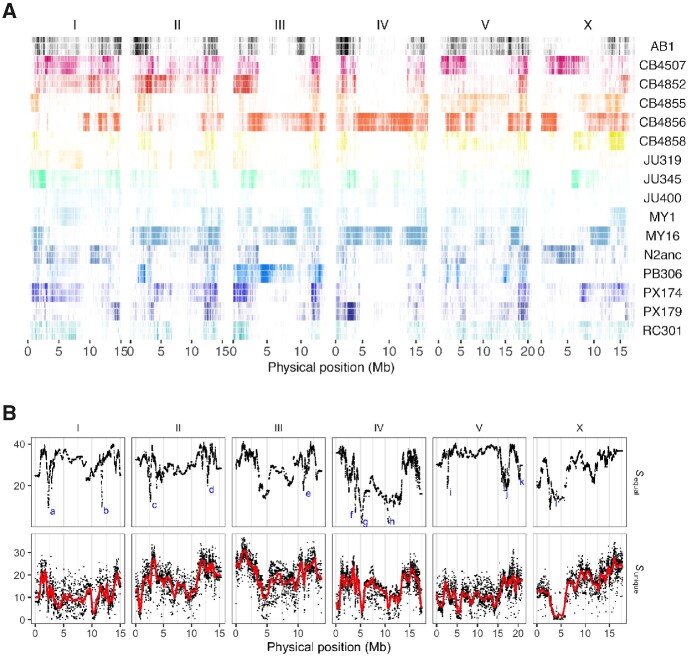
Figure 5.
(A) Founder representation across CeMEE populations. For each of the 16 founders, the composition of reconstructed haplotypes is shown for A6140 (top), CA[1-3]50 (middle) and CA[1-3]100 (bottom) RILs, averaging over populations for CA lines. Identity at segregating markers in 5 kb windows is plotted, color intensity scales with frequency (see Supplementary Figure S5A for quantities). (B) Upper row: haplotype divergence across all CeMEE RILs in each 5 kb window (entropy, high values indicating low divergence), against the expectation of equal representation of all 16 founders. Outliers highlighted span known loci zeel-1/peel-1 (a), fog-2 (k; an experimental artifact from introgression of a null allele into GT populations), and npr-1 (l). Other labeled, well-localized peaks of divergence correspond to selection of (b) CB4856 haplotypes upstream of vab-10, (c) hyperdivergent CB4852 haplotypes, (d) hyperdivergent MY16 haplotypes, centered on gsy-1, (e) MY1 haplotypes spanning vab-7, (f-h) long, recombined MY16 and CB4856 haplotypes nearing fixation, (i) hyperdivergent CB4856 haplotypes, and (j) MY16 haplotypes with unique variants in srr-3, cpr-2 and four other genes. Second row: as above, but against the expectation of equal proportions of the unique SNV founder haplotypes observed in each window, with a locally-weighted (LOESS) polynomial regression.