Fig. 1.
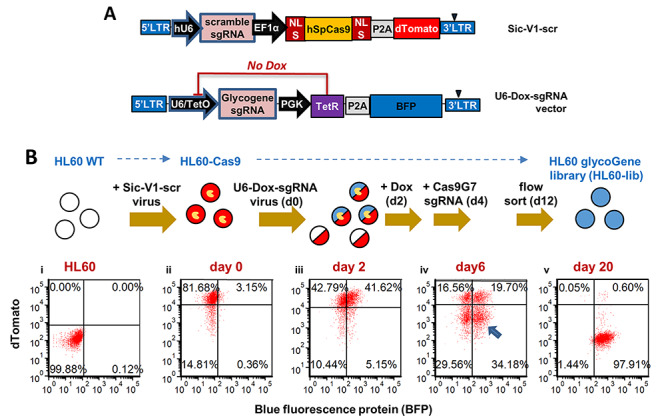
Creation of GlycoGene CRISPR library transduced cells. A. Schematic of lentiviral vectors used in the CRISPR screen. dTomato acts as a surrogate reporter of Cas9 nuclease activity in SiC-V1-scr. GlycoGene CRISPR library was cloned in a doxycycline-inducible vector with BFP fluorescence reporter. B. An HL60-Cas9 cell line was established by transduction with SiC-V1-scr vector, and single cell flow sorting for isogenic clone expressing high levels of dTomato reporter (Bi–ii). A virus pool synthesized from the glycoGene CRISPR sgRNA library (U6-Dox-sgRNA) was applied to these cells. Two days later, 42% of the cells were BFP positive (Biii). One microgram per milliliter Dox was added starting at day 2 to initiate gene editing. Dox was removed and Cas9G7 sgRNA electroporated on day 4 to inactivate Cas9 activity. dTomato−BFP+ cells (arrow) were FACS sorted at day 12 in order to establish the stable HL-60 glycoGene cell library (called “HL60-lib”). Flow cytometry plots represent: i. wild-type HL60; ii. HL60-Cas9; iii. HL60-Cas9 with glycoGene library; iv. cells after Cas9 nuclease editing on day 6; and v. HL60-lib cells on day 20. dTomato fluorescence of HL60-lib on day 20 (Bv) is similar to wild-type HL60s (Bi) confirming complete knockdown of Cas9 activity. (This figure is available in black and white in print and in color at Glycobiology online)
