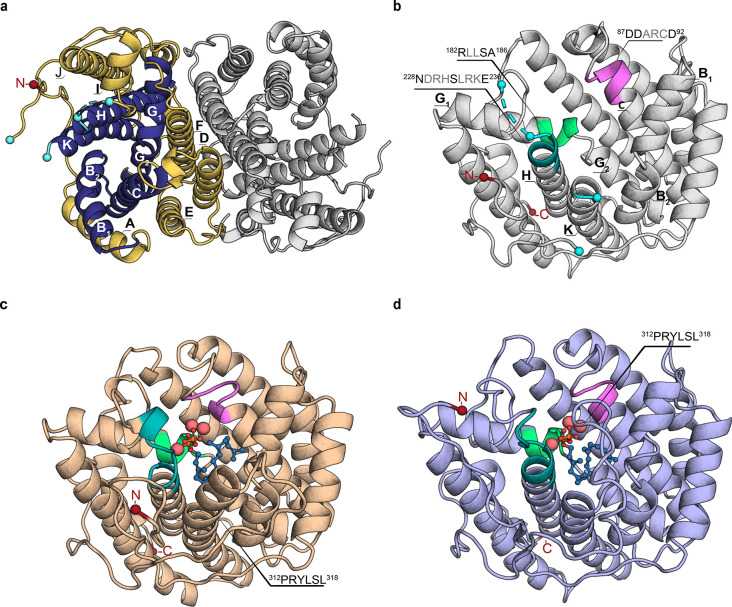
Figure 1.
Crystal structure of SvS-A2 and homology model of SvS-WT. (a) Top view of full dimeric crystal structure of reconstructed ancestral spiroviolene synthase SvS-A2, PDB-ID: 6TBD (2.30 Å). In one monomer, helices B, C, G, H, and K forming the active site cavity are shown in dark blue, peripheral helices A, D, E, F, I, and J are shown in gold. The first resolved N-terminal residue (Asp09) is shown as a red sphere. Residues that are flanking unresolved loops (Leu233/His241 and Arg311/Pro329) are shown as cyan spheres. (b) Front view of a monomer of the SvS-A2 crystal structure. The DDxx(x)D motif is shown in violet, NSE motif in teal, effector motif in light green. (c) Final model of SvS-A2 with missing loops and the trimetal ion cluster (shown as enlarged pink spheres) modeled and GGPP substrate (shown as blue sticks and spheres) docked as described in the Supporting Methods section. (d) Substrate-docked homology model of extant SvS-WT (monomeric) based on a surface variant of SvS-A2 (sequence-identity 78%). Motifs and additional molecules are shown as described in (c). The modeled C-terminal end of helix K (312PRYLSL318) shows considerable structural changes between SvS-A2 and SvS-WT.