Fig. 4.
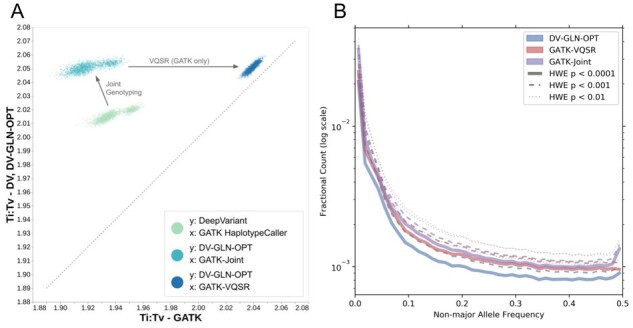
1KGP cohort callset quality. (A) Ti:Tv ratios of 1KGP samples, from single-sample SNPs and joint-called SNPs, generated by DV-GLN-OPT and GATK pipeline. Each point represents the ratio in one of the 2504 samples across the whole genome. Each point cloud compares the Ti:Tv ratios in variant calls from the two systems, after equivalent steps are performed. The first cloud (in light green) compares the Ti:Tv ratios from DeepVariant (y-axis) and GATK HaplotypeCaller (x-axis) single sample calls. The second cloud (in turquoise) compares Ti:Tv after joint-genotyping is performed (optimized GLnexus for DeepVariant, and GenomicsDBImport+GenotypeGVCFs for GATK HaplotypeCaller). Finally the third cloud (in blue) compares the final outputs from the two systems, after VQSR is performed for GATK (x-axis), while no additional operation is performed for the optimized DeepVariant-GLnexus calls. (B) Fractional counts of autosomal variants with low HWE p-values, binned by non-major allele frequency in DV-GLN-OPT, GATK-VQSR and GATK-Joint. The major allele is the allele with the largest allele count in a given variant within the callset. The variants are aggregated in non-major-allele-frequency bins of size 0.0125, and the frequency is clipped at 0.5 for visualization purposes (for all methods the fractional counts in bins after 0.5 are less than )
