Fig. 7.
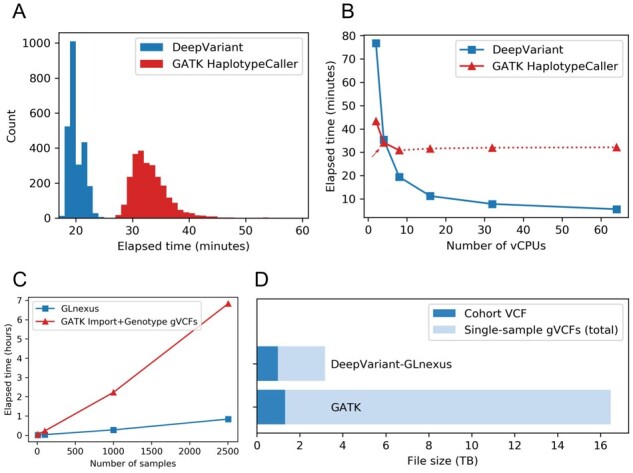
Cost benchmarking DeepVariant-GLnexus and GATK pipeline. (A) Distribution of elapsed real times to generate single-sample gVCF (chr22 only) from aligned reads across n = 2504 1KGP samples, using DeepVariant and GATK HaplotypeCaller (BQSR not included) in 8-vCPU machine. GPU/TPU acceleration was not used for DeepVariant. (B) Elapsed real times to generate gVCF (chr22 only) of one sample (NA12878) using a cloud machine with varying number of vCPUs, with DeepVariant and GATK HaplotypeCaller (excluding BQSR). The default value for HaplotypeCaller’s HMM multithreading flag (–native-pair-hmm-threads) is 4 (red arrow) and it was practically ineffectual for 16 vCPUs and more (red dotted lines). (C) Elapsed real times to merge the chr22 gVCF files from (A) into a cohort VCF for nested subsets of the 1KGP samples, using GLnexus (for DeepVariant gVCFs) and GATK GenomicsDBImport + GenotypeGVCFs (for HaplotypeCaller gVCFs). GATK VQSR step was not included. (D) The file sizes of the whole-genome cohort VCFs and the single-sample gVCFs of 1KGP samples from DeepVariant-GLnexus and GATK pipeline
