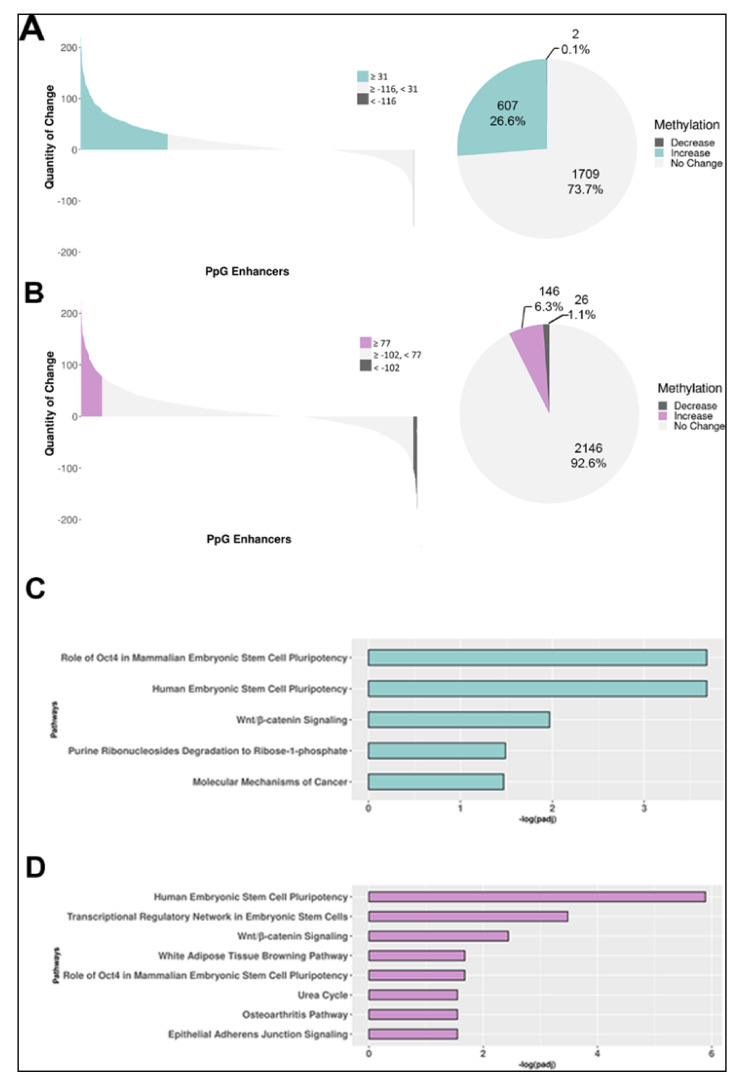
Figure 3.
DNA methylation change in LSD1-bound pluripotency gene enhancers during ESC differentiation. (A,B) Genome-wide DNA methylation analysis by MethylRAD sequencing. The number of MethylRAD reads per LSD1-bound pluripotency gene enhancer (PpGe) region were used as a measure for the extent of DNA methylation and compared between day 7 (D7) differentiated (untreated or treated) and undifferentiated (UD) samples. Untreated (A); treated (B). The left panel of the waterfall plot shows DNA methylation changes in pluripotency gene enhancers (PpGes), which were computed by subtracting normalized read counts in D7 differentiated samples from normalized counts in UD samples. Upper and lower quartiles were used in thresholding regions as gaining or losing methylation. The right panel of the pie chart shows fractional distribution of PpGes with an increase, decrease, or no change in DNA methylation. (C,D) Statistically significant enriched canonical pathways among the genes associated with increased DNA methylation in enhancers. (C) Pathways that had higher enhancer DNA methylation in D7 untreated differentiated samples than UD. (D) Pathways that had higher enhancer DNA methylation in pargyline-treated D7 differentiated samples than in UD. The x-axis shows the log10 (adjusted p-value), with the p-values adjusted for multiple testing using the Benjamini–Hochberg method.