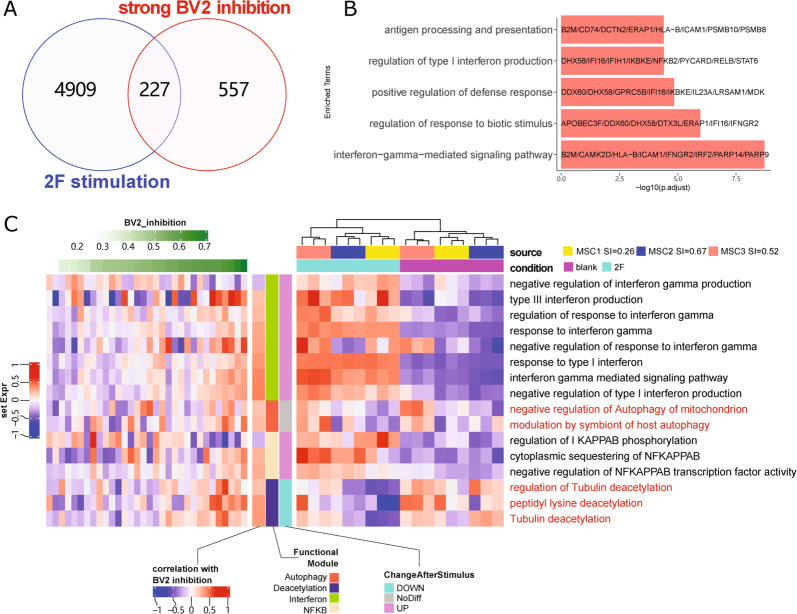
Fig. 6. Two transcriptomic data sets (32 huMSC lines and 3 huMSC lines with and without 2-factor stimulation) revealed common genes and pathways.
A Venn diagram shows the overlap between genes up-regulated by 2-factor stimulation and genes positively correlated to BV2 inhibition capacity. B GO enrichment analysis of overlapped 227 genes. C GSVA heatmap shows specific GO-term associated gene sets in 32 huMSCs that positively or negatively correlated with BV2-cell immune suppressive capacity, and their expression levels before and after 2-factor stimulation. Results clearly indicated that interferon and NF-κB pathway associated genes were upregulated upon 2-factor stimulation, whereas genes related to autophagy and protein deacetylation did not correlate well with 2-factor stimulation.