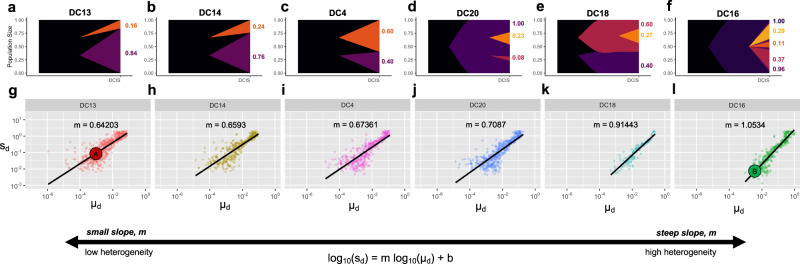
Fig. 3. Model parameterization of DCIS heterogeneity.
a–f Muller diagrams of inferred clonal heterogeneity of the six polyclonal synchronous DCIS-IDC patients from Casasent et al.20, arranged from low (a) to high (f) heterogeneity. g–l The mathematical model is extended to a three-dimensional domain and constrained to grow inside a ductal network (Fig. 4a) and 10,000 stochastic simulations for a range of driver mutation rates (μd ∈ [10−7, 1]) and fitness (sd ∈ [10−3, 10]) are simulated to N = 104 cells. Parameterizations for which clonal diversity of the mathematical model (eqn. (3)) lie within error bounds reported in Casasent et al. are plotted and fit to eqn. (4). As heterogeneity increases in a–f, the slope of this best fit (m) also increases in g–l. Source data for a–f are provided as a Source Data file.