Figure 5.
Development of a novel peptide that disrupts the interaction between SLP76 and ITK
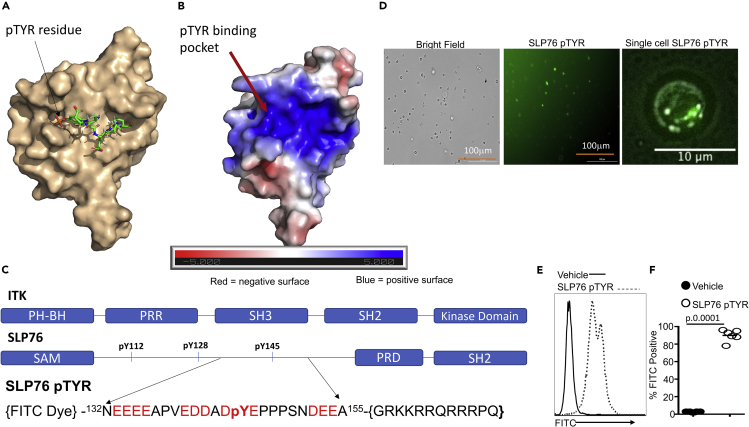
(A) NMR spectroscopy structure of murine ITK SH2 domain showing its complex with a peptide containing a pTyr residue (PDB code:2ETZ) that was previously solved by Pletneva et al. (46). The SH2 domain is rendered in surface representation (wheat), while the peptide derived from residues 143–148 of SLP76 with a sequence (143ADpYEPP148) is shown in stick model. In contrast, our SLP76pTYR inhibitor consists of residues 132–155 of SLP76.
(B) Electrostatic profile is shown, calculated using the APBS plugin in Pymol.
(C) Top: Organization of the domain architecture of full-length ITK showing the c-terminal Kinase domain, Src-homology 2 (SH2), the Src-homology 3 (SH3) domains, the intrinsically disordered proline-rich region (PRR), and the N-termimal Pleckstrin homology (PH) and Tec homology (TH) domains. Bottom: Organization of the domain architecture of full-length SLP76 adaptor protein showing the N-terminal SAM domain, the intrinsically disordered region containing phosphotyrosines pY112, pY128, pY145, which are followed by a proline-rich domain (PRD) and a C-terminal SH2 domain. Bottom: Design of the novel peptide, SLP76pTYR with an N-terminal FITC to monitor the peptide in cells and a C-terminal TAT sequence (GRKKRRQRRRPQ TAT sequences) for cell membrane permeability. Also shown are the amino acid sequence of residues 132–155 of SLP76, which was used to design the SLP76pTYR peptide inhibitor.
(D) T cells were examined for percentage FITC positive by fluorescence microscopy. A single cell in focal plane near the cover glass was imaged.
(E) Primary cells cultured with SLP76pTYR or the non-specific peptide were washed and examined for FITC expression by flow cytometry.
(F) Quantification of the FITC expression for (E). We used two-way ANOVA for statistical analysis and confirmed our statistical finding by Student's t-test was performed. See also Figure S7.