Fig. 4.
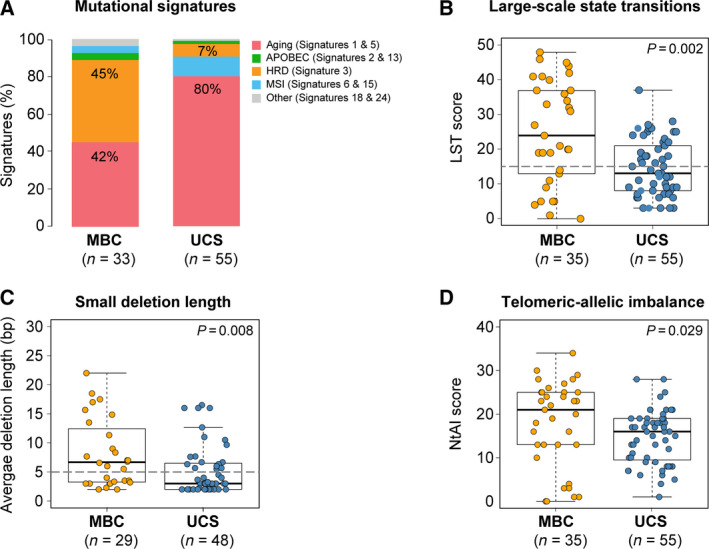
Genomic features of homologous recombination repair deficiency in metaplastic breast carcinomas and uterine carcinosarcomas. (A) Mutational signatures in metaplastic breast cancers (MBCs) from Ng et al. [2] and nonhypermutated uterine carcinosarcomas (UCSs) from TCGA [19] identified using DeconstructSigs [39]. Mutational signatures are color‐coded according to the legend and were only performed for samples ≥ 20 SNVs. (B) LST scores in MBCs from Ng et al. [2] and nonhypermutated UCSs from TCGA [19]. The gray line depicts the cutoff for LST high (≥ 15) [36]. (C) Small deletion length in MBCs from Ng et al. [2] and nonhypermutated UCSs from TCGA [19] according to Alexandrov et al. [40], which in HRD‐defective tumors has been found to be ≥ 5 nucleotides (gray line). (D) NtAI score in MBCs from Ng et al. [2] and nonhypermutated UCSs from TCGA [19] according to Morganella et al. [41]. Mann–Whitney U‐test was performed for comparisons in (B), (C), and (D). MBC, metaplastic breast cancer.
