Fig. 2.
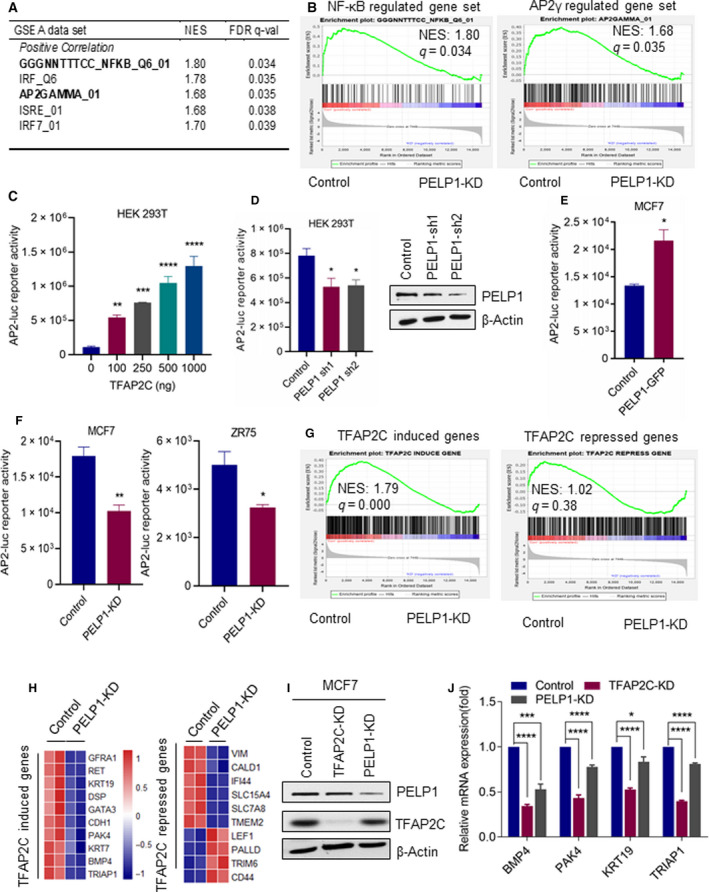
PELP1 serves as coactivator of TFAP2C. (A) Top TFs enriched in PELP1‐regulated genes identified using published PELP1 RNA‐seq data. (B) GSEA of target genes of NF‐κB and TFAP2C with PELP1 RNA‐seq data. (C) HEK293T cells were transfected with AP‐2 Luc plasmid along with increasing concentrations of TFAP2C, and 48h after transfection, the reporter activity was measured. (D) HEK293T cells stably expressing either control or PELP1 shRNA were cotransfected with AP‐2 Luc plasmid along with TFAP2C (100 ng), and 48 h after transfection, the reporter activity was measured. PELP1 knockdown was confirmed using western blotting. (E) MCF7 control and MCF7‐GFP‐PELP1 cells were transfected with AP‐2 Luc plasmid, and the reporter activity was measured after 48 h. (F) MCF7 control and MCF7‐PELP1 shRNA cells or ZR75‐control and ZR75‐PELP1 shRNA cells were transfected with AP‐2 Luc plasmid, and the reporter activity was measured after 48 h. (G) GSEA testing correlation of target genes of PELP1 with genes upregulated by TFAP2C and repressed by TFAP2C. (H) Heat map showing changes in expression levels of several TFAP2C‐upregulated and TFAP2C‐repressed genes in PELP1 RNA‐seq data. (I) TFAP2C knockdown was confirmed using western blotting. (J) Total RNA was isolated from MCF7 model cells expressing either PELP1 shRNA or TFAP2C shRNA, and the status of TFAP2C‐upregulated genes was validated using RT–qPCR. Data are shown as the means ± SEM of three experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by Student'st‐test in C–F and J.
