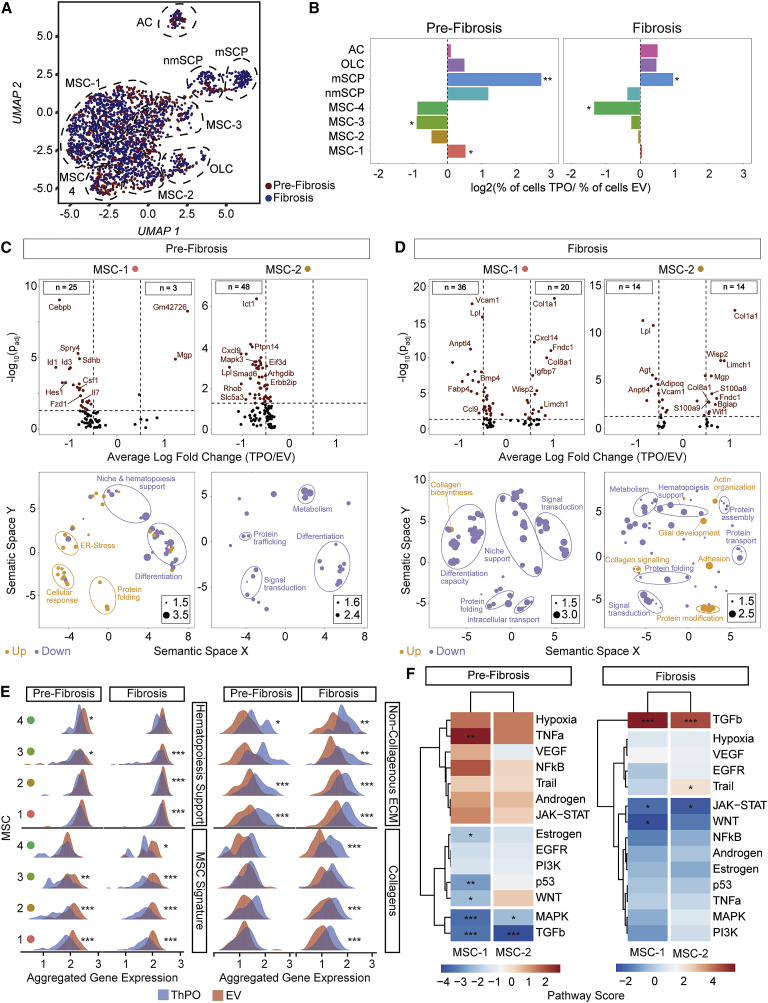
Figure 3.
Mesenchymal stromal cells −1 and −2 are reprogrammed in pre-fibrosis and acquire a pro-fibrotic phenotype in fibrosis
(A) UMAP of non-hematopoietic single cells in pre-fibrosis (5 weeks; red) and fibrosis (10 weeks; blue).
(B) Bar plot of cells per cluster in ThPO versus EV after normalization to input per condition. Fisher’s exact test with Bonferroni correction was used.
(C and D) Volcano plot (ThPO versus EV) of differentially expressed genes (Wilcoxon rank-sum test, two tailed; calculated per cluster individually; top panel). Associated gene ontology terms (biological processes; BP; Fisher exact test; single tailed) plotted semantically (bottom panel) in (C) pre-fibrosis and (D) fibrosis. (For a complete list of GO-terms and corresponding genes, see Tables S2 and S3.)
(E) Ridgeline plot of functional gene sets (as indicated) in ThPO (blue) versus EV (red). Competitive gene set enrichment analysis.
(F) PROGENy analysis of MSC-1 and MSC-2 in pre-fibrosis and fibrosis. Sampling based permutation (10,000 permutations). Pathway activity scores as Z scores.