Figure 4.
Mesenchymal stromal cells −1 and −2 are reprogrammed in JAK2V617F-induced primary myelofibrosis
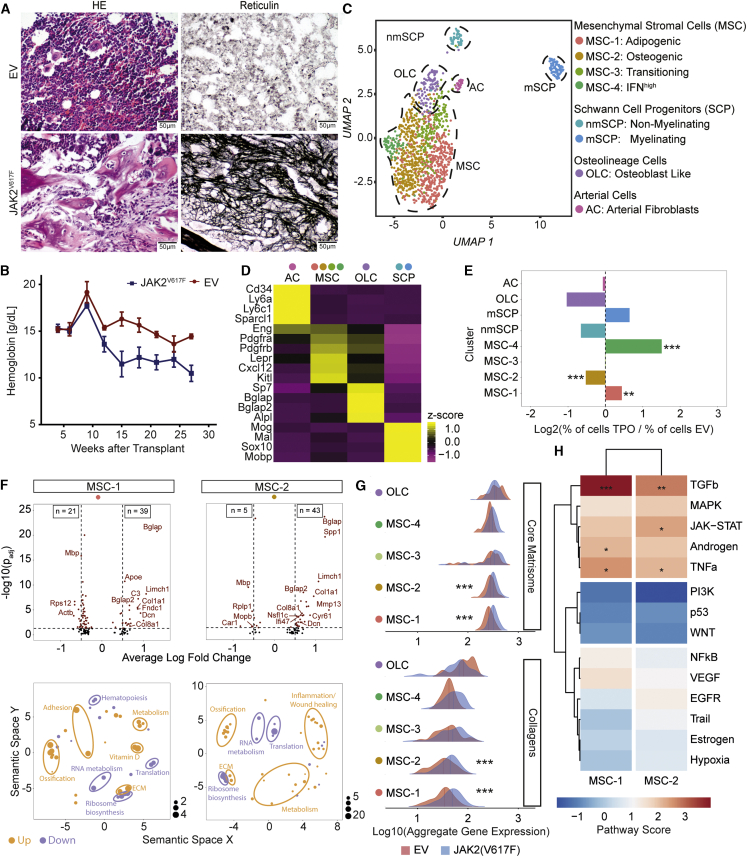
(A) Representative HE and reticulin staining of JAK2V617F-induced fibrosis or control (EV, n = 4 mice per condition). Scale bar, 50 μm.
(B) Hemoglobin levels (mean ± SEM). Two-way ANOVA with post hoc pairwise t test was used.
(C) UMAP of non-hematopoietic BM cells (n = 1,292 cells).
(D) Top marker genes. Wilcoxon rank-sum test, p < 0.01. A full list of marker genes and corresponding GO terms is provided in Table S2.
(E) Bar plot of cells number in JAK2V617F versus control (EV) after normalization to total number. Fisher’s exact test with Bonferroni correction was used.
(F) Volcano plot; Wilcoxon rank-sum test, two tailed; calculated per cluster between JAK2V617F and JAK2WT (top panel). Associated gene ontology terms (biological processes; BP) plotted semantically (bottom panel) are shown. (For a complete list of GO-terms and corresponding genes, see Table S3.)
(G) Ridgeline plot for matrisome gene sets in MSC and OLC clusters (JAK2V617F; blue versus control; red) condition. Competitive gene set enrichment analysis was used.
(H) PROGENy analysis of MSC-1 and MSC-2. Sampling based permutation (10,000 permutations). Pathway activity scores as Z scores are shown.