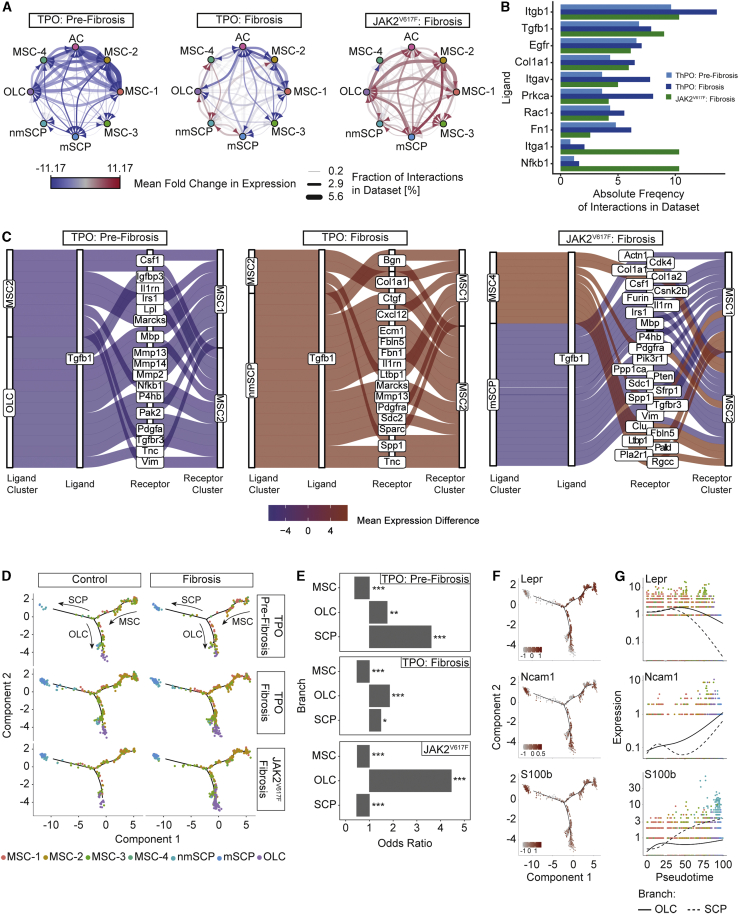
Figure 5.
Tgfb1-driven cellular interaction and biased differentiation of MSCs in MPN
(A) Network plot of ligand-receptor activity in control and fibrosis.
(B) Bar plot of top 10 most abundant ligands in all inferred ligand-receptor interactions per dataset. Datasets are shown side by side (ThPO: pre-fibrosis, light blue; ThPO: fibrosis, dark blue; JAK2V617F: fibrosis, green).
(C) Top 20 deregulated interactions mediated by Tgfb1 targeting MSC-1 and MSC-2. Interactions ordered based on difference in mean LR expression between fibrosis and control. (For a full list of inferred ligand-receptor interactions, see Table S5.)
(D) Reconstructed cell differentiation trajectory of MSC populations.
(E) Bar plot of numerical changes between fibrosis and control in respective branches as identified in pseudotime analysis. Fisher’s exact test with Bonferroni correction was used.
(F) Expression levels of highlighted genes projected onto differentiation trajectory in pseudotime space.
(G) Expression levels of indicated genes with respect to their pseudotime coordinates. Black line indicates differentiation trajectory toward OLC branch and dotted lines toward SCP branch.