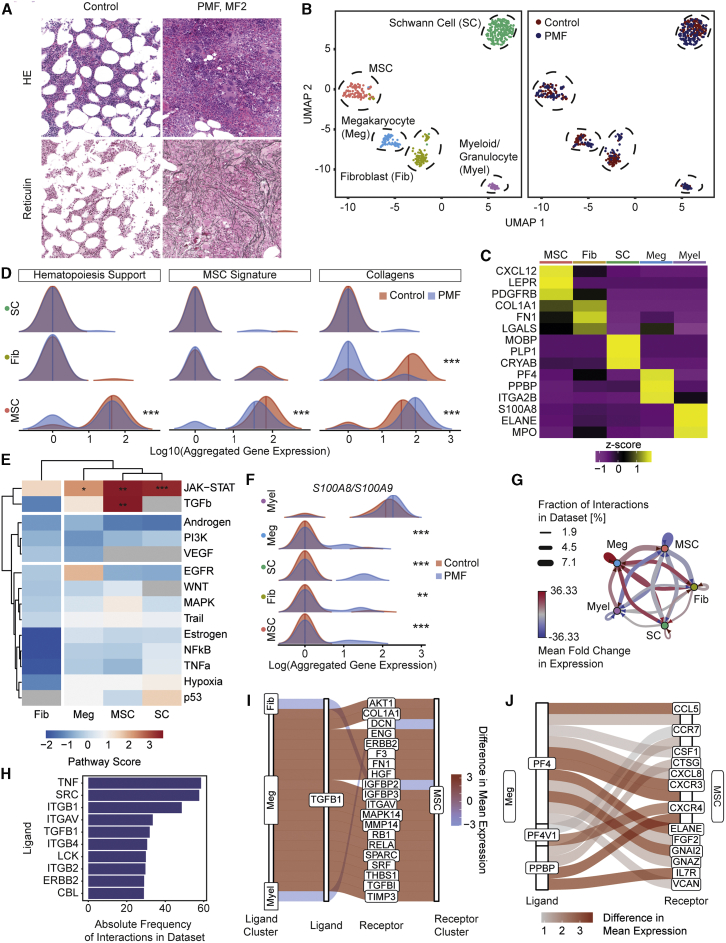
Figure 6.
MSCs are fibrosis-driving cells in patients characterized by upregulation of S100A8/A9
(A) Diagnostic BM images of the patients. Representative H&E and reticulin stainings. For additional images (all controls) and detailed patient characteristics, see Figure S5.
(B) UMAP of cells in 1 PMF patient (MF2, n = 243 cells) and two control patients (MF0, n = 255 cells). In the left panel, cells are color coded by their annotated cellular identity, and in the right panel, by their patient source.
(C) Top marker genes. Wilcoxon rank-sum test, p < 0.01.
(D) Ridgeline plot comparing PMF (blue) versus control (red) condition. Competitive gene set enrichment analysis was used.
(E) PROGENy analysis. Sampling-based permutation (10,000 permutations). Pathway activity scores are given as Z scores.
(F) Ridgeline plot of S100A8/A9 expression in PMF (blue) or control (red). Significance estimated by modeling the dropout rate as a binomial process with the observed dropout rate per condition as estimator of p for both conditions, respectively.
(G) Network plot of ligand-receptor activity in PMF compared to control.
(H) Bar plot of top 10 most abundant ligands in all inferred ligand-receptor interactions.
(I) Sankey plot of top 20 deregulated TGFB1-mediated ligand-receptor interactions. The absolute difference in mean LR expression was used as a metric for the extent of deregulation.
(J) Sankey plot of top 20 deregulated ligand-receptor interactions mediated by PF4, PF4V1, or PPBP. The absolute difference in mean LR expression was used as a metric for the extent of deregulation.