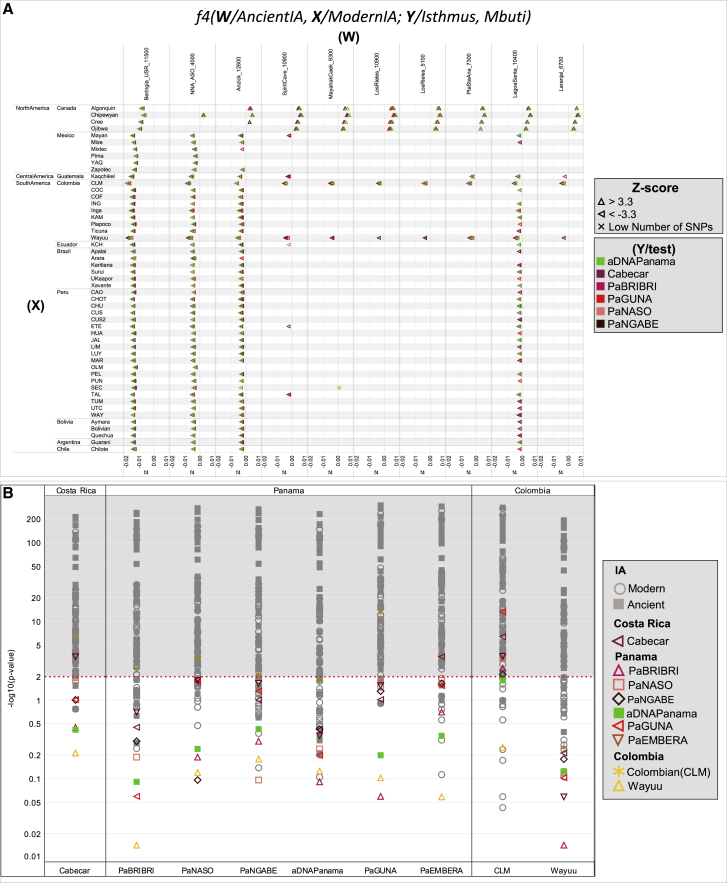
Figure 6.
f4 statistic tests on Isthmian and other IA groups and minimum number of ancestral sources
(A) f4 statistics in the form f4 (W/Ancient IA, X/Modern IA; Y/Isthmus, Mbuti) on uIA89, mIA417, and ancient genomes considering only transversions. Only sub-groups of meaningful ancient genomes were considered (see Table S4 for comparisons with the entire ancient dataset). The Emberá group was excluded due to a significant degree of admixture detected in the individuals. Each tested population (Y) is shown (with triangles pointing to X or W population) only when the initial conformation of the tree is rejected (p ~ 0.001, when the Z score is >|3.3|), thus visually pointing to the closer population (W or X) in each comparison.
(B) We used qpWave to compare (in pairs) ancient Panama and present-day Isthmian groups with all IA populations (considering rank 1). Outgroups were kept to the minimum and chosen to represent different IA ancestries identified here (STAR Methods) and in other papers. Rank 1 refers to a model in which all paired populations fit as derived from two ancestral sources, relative to the outgroups. A p value > 0.01 (< 2 in −log10 scale, dotted red line) means pairs that could be explained by a single independent source.