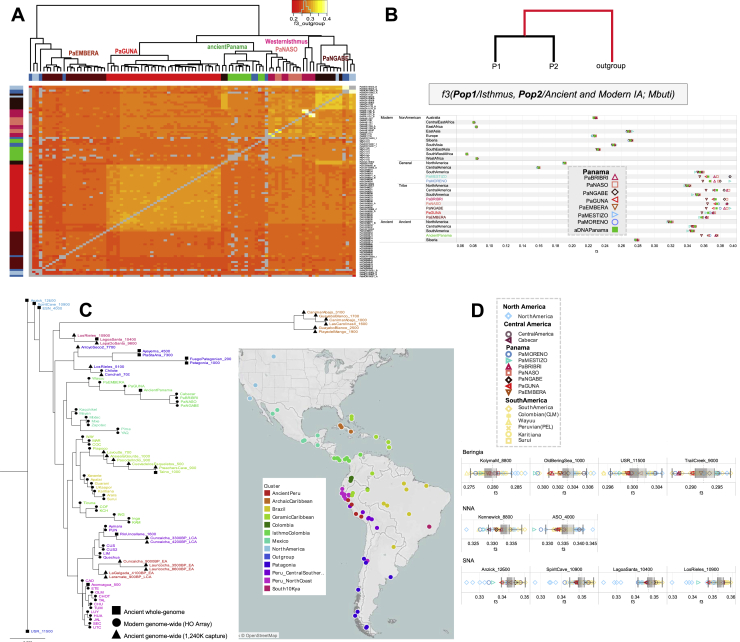
Figure S3.
f3 statistics involving Isthmian individuals, related to Figures 2, 3, and 6
(A) Heatmap based on outgroup f3-statistics. The shared drift among the Panamanian individuals was analyzed considering those included in uIA217 plus ancient individuals and masked data in mIA417. Color intensity is inversely proportional to the shared ancestry among individuals, which was used to build the dendrogram. (B) Outgroup f3 statistics where ancient and modern Isthmian groups (Pop1) were compared to worldwide populations (Pop2) including non-American groups in the rWD1560 dataset, all populations in the mIA417 and uIA217 datasets and all ancient individuals. All comparisons have a Z score > 32.912. The average f3 value for each population is reported in abscissa. (C) The neighbor-joining tree is built using the inverse values derived from the outgroup f3 statistics on all Central and South American populations pairs plus Anzick-1, Early San Nicolas (ESN), Spirit Cave and USR-1. The latter is considered as an outgroup in the tree. We retained only populations with more than 30K overlapping SNPs and significant Z scores (> 3.3) in all comparisons. The map shows the geographic distribution of the populations, which are colored according to their genetic proximity in the tree. (D) We also analyzed the shared genetic history of modern IA populations (included in mIA417 and uIA89) against ancient reference genomes from Beringia and the Americas (representative of the NNA and SNA ancestries). Boxplots in gray help to visualize the distribution of f3 values in each comparison, indicating the median (most typical) value, 25th and 75th percentiles (dark and light gray, respectively), and arms extending 1.5 times the IQR (interquartile range). The dotted line indicates the f3 average value.