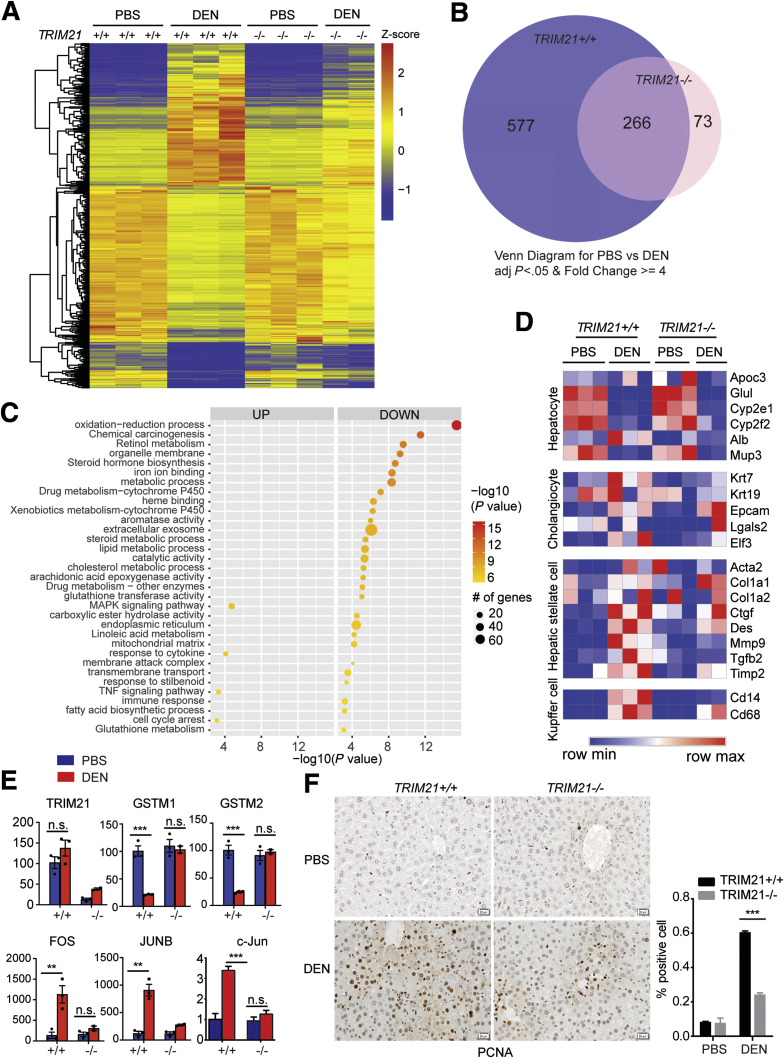
Figure 5.
DEN-induced hepatocyte proliferation is suppressed in TRIM21–/–liver. (A) Eight-week-old TRIM21+/+ and TRIM21–/– male mice were treated with PBS or 200-mg/kg DEN by intraperitoneal injection. Liver tissues were collected after 48 hours for RNA-seq. Hierarchical clustering and heatmap illustration of differentially expressed genes under indicated conditions. Color key indicates Z-scores after normalization. (B) Venn diagram showing overlap of genes differentially expressed in PBS-treated vs DEN-treated TRIM21+/+ and TRIM21–/– mice. Analysis was restricted to genes, which showed a 4-fold difference and yielded P < .05. (C) Dot plots showing the enriched terms from Gene Ontology analysis using differentially expressed genes identified from DEN vs PBS treatment in TRIM21 WT mice (the 577 unique genes altered by >4-fold in TRIM21-WT mice as shown in B). Note that genes involved in redox regulation, chemical carcinogenesis, and retinol metabolism are identified. (D) Heatmap shows that DEN-induced expressions of hepatic stellate cell and Kupffer cell markers were markedly decreased in TRIM21 KO livers, while no obvious difference was detected in hepatocyte markers. (E) Quantitative reverse-transcription PCR for the DEN-treated TRIM21 WT and KO liver samples, showing decreased antioxidant response and increased proliferation signals DEN-treated WT mice (n = 3 per group). (F) IHC analysis of PCNA expression in livers of PBS- and DEN-treated TRIM21 WT and KO mice. PCNA-nuclear positive cells were counted and the ratio of positive cells was calculated. Representative images are shown. Data are presented as means ± SEM of 3 mice in each group. ∗∗∗P < .001. FPKM, fragments per kilobase of transcript per million; n.s., nonsignificant.