Figure 1. The ubiquitin modification system.
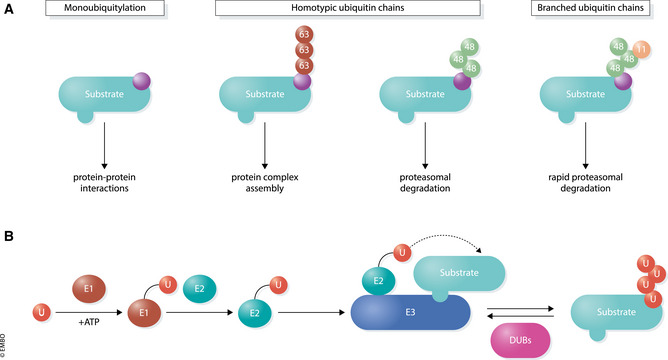
(A) Various structures and functions of different types of ubiquitin conjugates. Monoubiquitylation (left) involves transfer of a single ubiquitin to a substrate. E3 ligases can also connect several ubiquitin molecules together using the C‐terminus of one subunit and one of seven lysine (K) residues (K6, K11, K27, K29, K33, K48, K63) or the N‐terminal methionine (M1) on the other. Different ubiquitin topologies adopt distinct structural conformations which direct a wide array of substrate outcomes. (B) Ubiquitylation occurs through an enzymatic cascade. E1 enzymes use ATP to form a high‐energy thioester bond to ubiquitin through an active site cysteine residue. Charged E1s transfer ubiquitin to one of ~ 40 E2 ubiquitin‐conjugating enzymes, which then bind to one of ~ 600 E3 ubiquitin ligases that facilitate the transfer of ubiquitin onto a specific substrate. Approximately 100 DUBs remove ubiquitin from substrates to reverse the ubiquitylation process.
