Figure 3. DDX43 unwinds Ago3‐cleaved RNA and drives Siwi‐piRISC biogenesis.
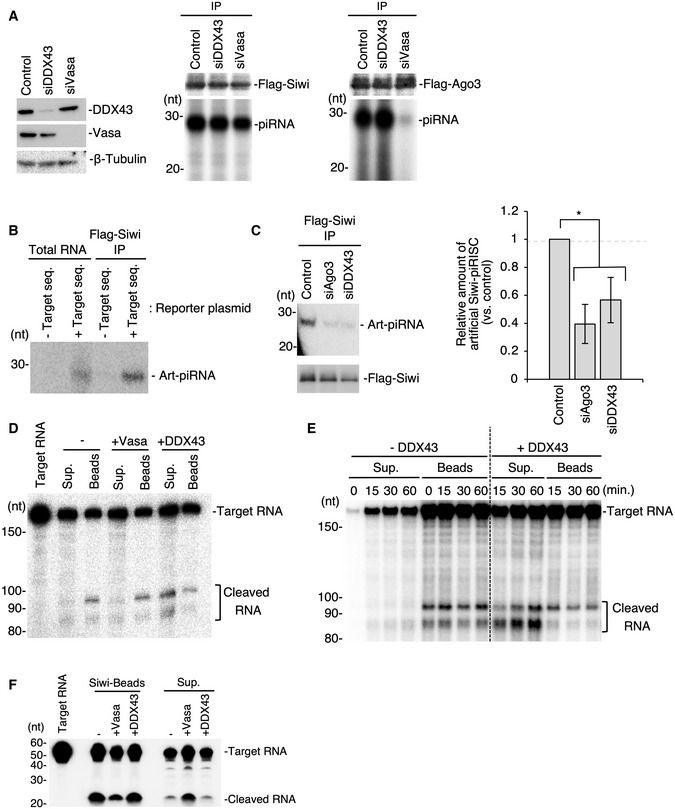
- Analysis of Siwi‐piRNA and Ago3‐piRNA production in DDX43‐ or Vasa‐knockdown BmN4 cells. Left: The expression levels of DDX43 and Vasa following RNAi treatment. Middle and right: Comparison of Siwi‐piRNA and Ago3‐piRNA levels following DDX43 or Vasa knockdown by RNAi. The piRNAs were isolated by the immunoprecipitation of Flag‐tagged proteins from knockdown cells transfected with Flag‐Siwi or Flag‐Ago3 and then 32P‐labeled.
- Artificial Siwi‐piRNA production from a reporter plasmid encoding the PiggyBac‐piRNA target sequence.
- Analysis of the artificial piRNA levels in DDX43‐knockdown BmN4 cells. Left: Artificial piRNA and Flag‐Siwi were detected by Northern blotting and Western blotting, respectively. Right: Quantification of artificial Siwi‐piRISC formed by the ping‐pong cycle pathway. Data represent the mean ± standard deviations, n = 3 independent experiments. *P < 0.05 as determined with the t‐test.
- RNA‐unwinding assays upon Ago3‐piRISC‐dependent target RNA cleavage (Fig EV3D). Recombinant Vasa or DDX43 was added and incubated in the reaction mixture upon cleavage and the materials were separated using a magnet to supernatant (Sup.) and bead (Beads) fractions. RNAs cleaved by Ago3 are shown as “Cleaved RNA”. DDX43 but not Vasa released the cleaved target RNA from Ago3‐piRISC.
- Time dependency of the DDX43 unwinding activity. Sup: supernatant.
- RNA‐unwinding assays upon Ago3‐piRISC‐dependent target RNA cleavage as in (D). Recombinant DDX43 was added and incubated in the reaction mixture upon cleavage for 0 min (i.e., no cleavage), 15, 30, and 60 min, and the materials were separated using a magnet to supernatant (Sup.) and bead (Beads) fractions. RNAs cleaved by Ago3 are shown as “Cleaved RNA”. DDX43 fails to unwind cleaved RNAs from Siwi‐piRISC. – DDX43 means no DDX43 added in the reaction mixture.
