Figure 5.
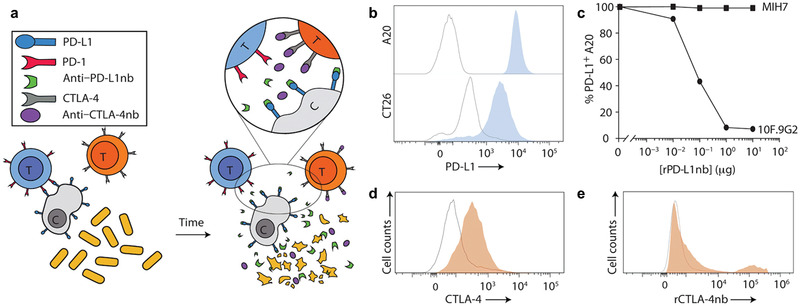
Engineering design and characterization of a bacteria‐based cancer treatment platform for in situ release of immune checkpoint inhibitor. a) Mechanism illustration by which engineered bacteria controllably and sustainably release PD‐L1 and CTLA‐4 nanobodies. b) Flow cytometric data of PD‐L1 expression on A20 and CT26 cells (outlined peaks, negative control; blue‐filled peaks, PD‐L1). c) Binding behavior of rPD‐L1nb to the 10F.9G2 and MIH7 PD‐L1 epitopes on A20 cells. d,e) Isolated splenocytes from naive C57BL/6 mice were analyzed by flow cytometry for d) intracellular CTLA‐4 expression (outlined peak, unstimulated CD3+ splenocytes; orange‐filled peak, PMA/ionomycin‐simulated CD3+ splenocytes) and e) rCTLA‐4nb binding to extracellular CTLA‐4 (outlined peak, secondary anti‐HIS antibody alone gated on CD3+ splenocytes; orange‐filled peak, rCTLA‐4nb gated on CD3+ splenocytes). Reproduced with permission.[ 46 ] Copyright 2020, The American Association for the Advancement of Science.
